Compositional Heatmaps
Example 1.1
We first import the packages used in this tutorial.
We also import Tengeler2020 from the mia package and store it into a variable.
Next, we transform the counts assay to relative abundance assay and store it into the TreeSE.
Then, we agglomerate the experiment to the order level, so that information is more condensed and therefore easier to visualise and interpret.
Why relative abundances?
Microbiome data is compositional. Relative abundance helps us draw less biased comparisons between samples.
Show code
# Import packages
library(miaViz)
library(patchwork)
# Plot composition by counts
counts_bar <- plotAbundance(tse_order, rank = "Phylum", use_relative = FALSE) +
ylab("Counts")
# Plot composition by relative abundance
relab_bar <- plotAbundance(tse_order, rank = "Phylum", use_relative = TRUE) +
ylab("Relative Abundance")
# Combine plots
(counts_bar | relab_bar) +
plot_layout(guides = "collect")
Figure 1: Sample composition by counts (left) or relative abundance (right).
Example 1.2
To reduce data skewness, we further transform the relative abundance assay with the Centered-Log Ratio (CLR), which is defined as follows:
\[ clr = log \frac{x}{g(x)} = log(x)−log[g(x)] \]
where x is a feature and g(x) is the geometric mean of all features in a sample.
Lastly, we get the row-wise z-scores of every feature from the clr assay to standardise abundances across samples.
Example 1.3
Finally, we visualise the clr-z assay with ComplexHeatmap.

Figure 2: Heatmap of CLR-Z assay where columns correspond to samples and rows to taxa agglomerated by order.
Why clr-z transformation?
A CLR-z transformation improves comparability in two steps:
- Apply CLR transform to center features column-wise
- Find Z score to standardise features row-wise
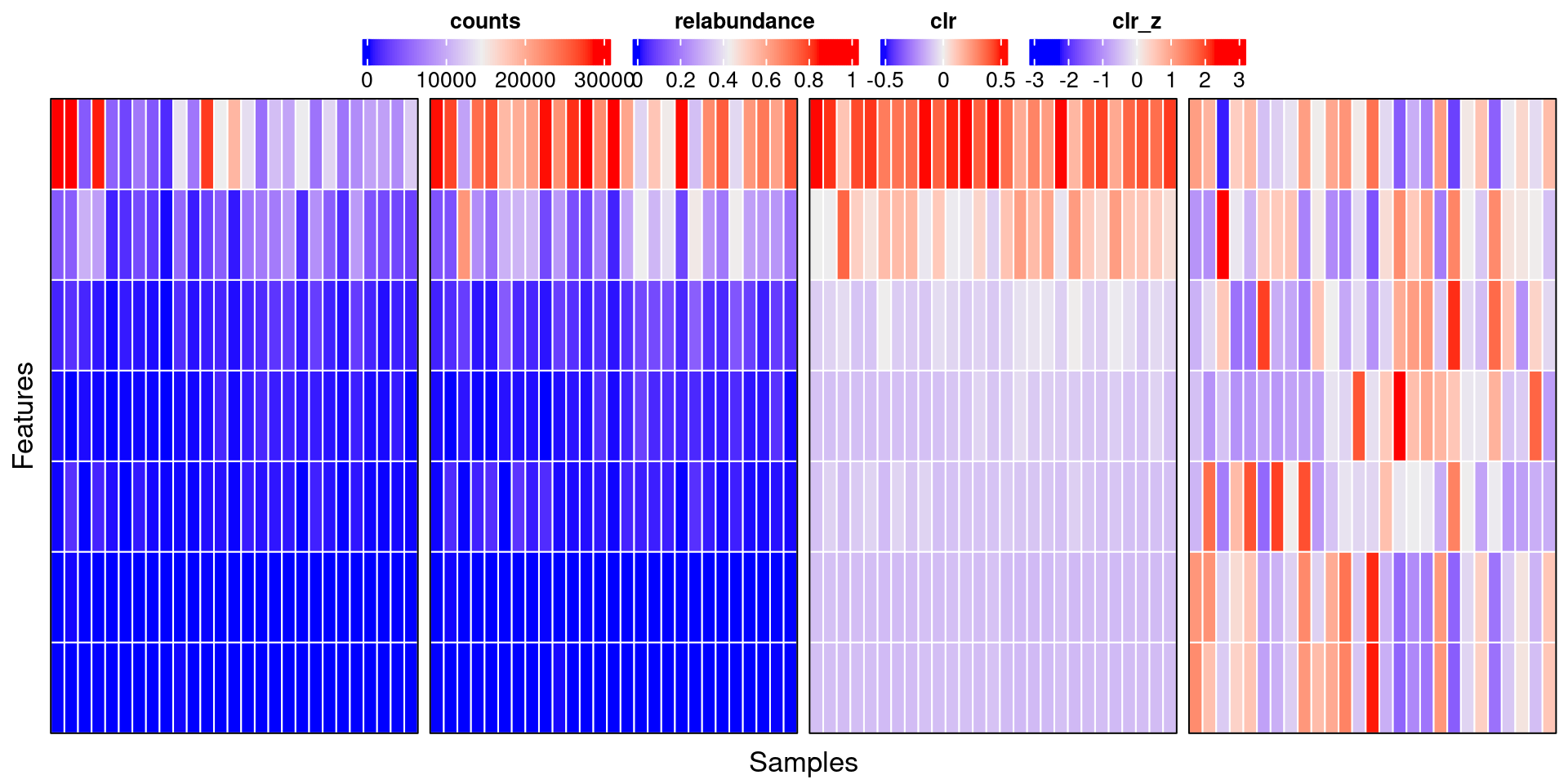
Figure 3: Visual comparison between counts, relative abundance, clr and clr-z assays (from left to right).
Exercise 1
- heatmap visualisation: exercise 9.2
Extra:
- advanced heatmap: exercise 9.3