Tabular data analysis
Recap of Day 1
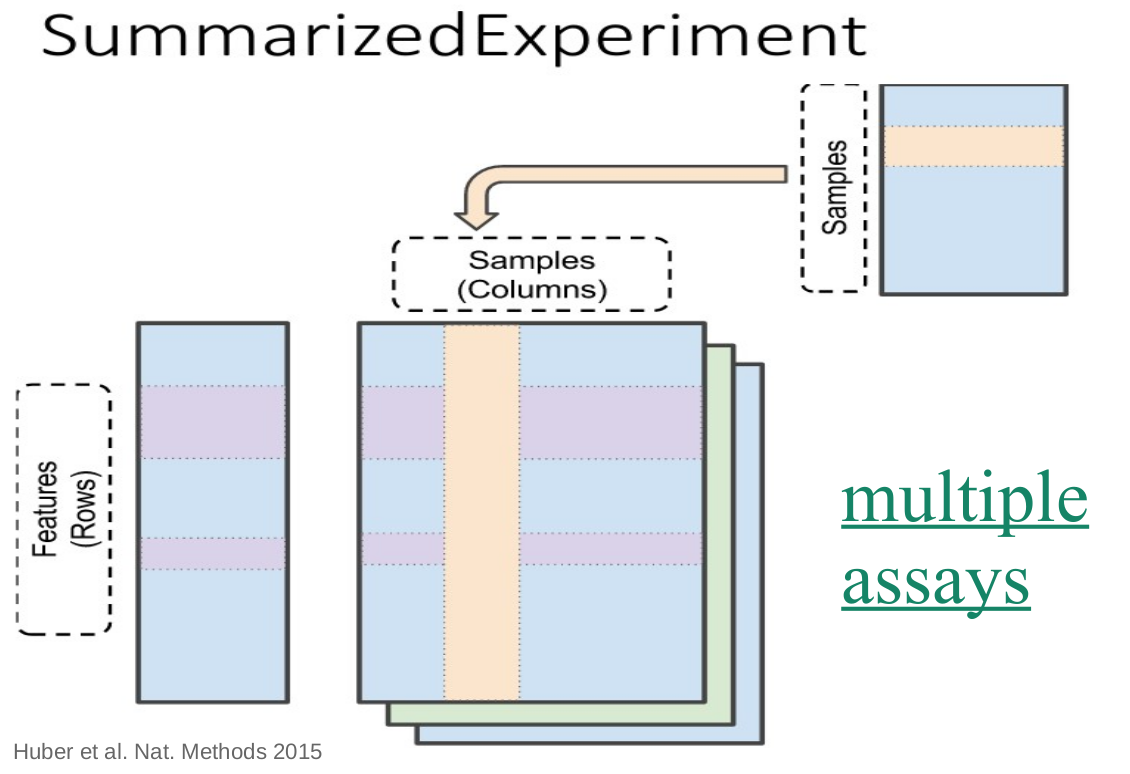
Day 1: Basic data wrangling
- reproducible data science workflow
- data import
- data containers
Today’s learning goals
- data manipulation (subsetting, transformations)
- augmenting the data (add diversities)
Today’s program
Morning: data wrangling
Afternoon: data visualizations
Data enrichment
Visualizing colData
Task: visualize the abundance of a specific microbial Species against the measurement Site
Alpha diversity task
Use the available tools to assess and visualize alpha diversity, and augment colData
- Exercises 17.5.1-17.5.2
- Add Shannon diversity in colData
- Visualize diversity differences between sample groups
Alpha diversity & aging
Healthy & normal obese subjects.
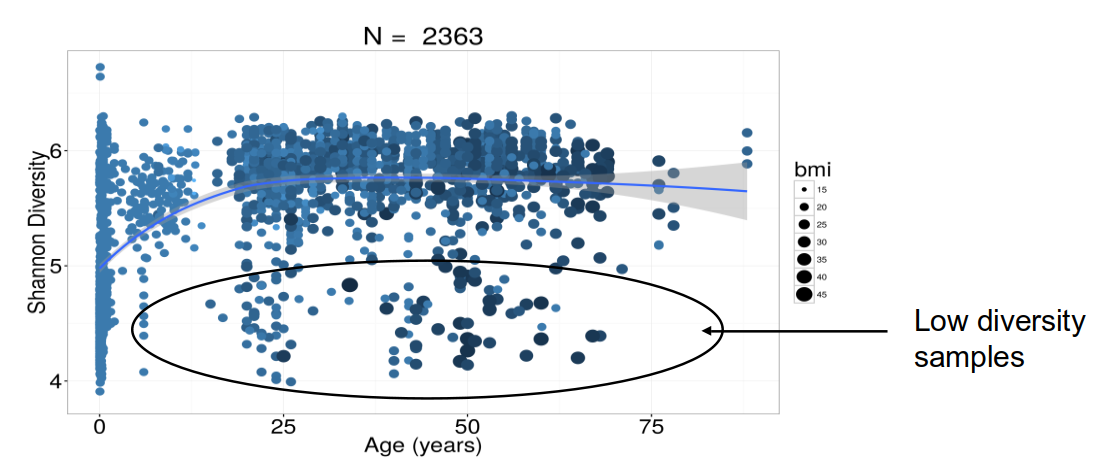
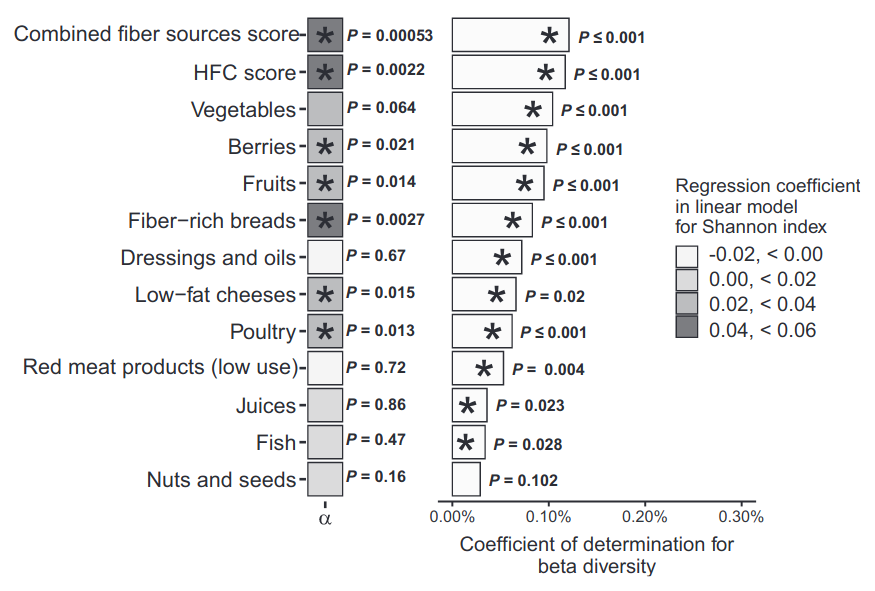
Alpha diversity and diet

Alpha diversity
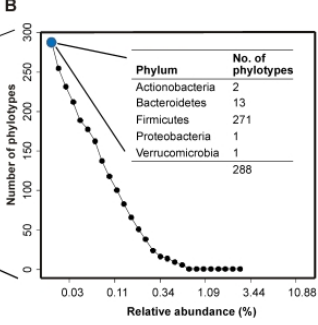
How many types?
Distribution of types?
Dominance of types?
Alpha diversity
How many types?
Distribution of types?
Dominance of types?
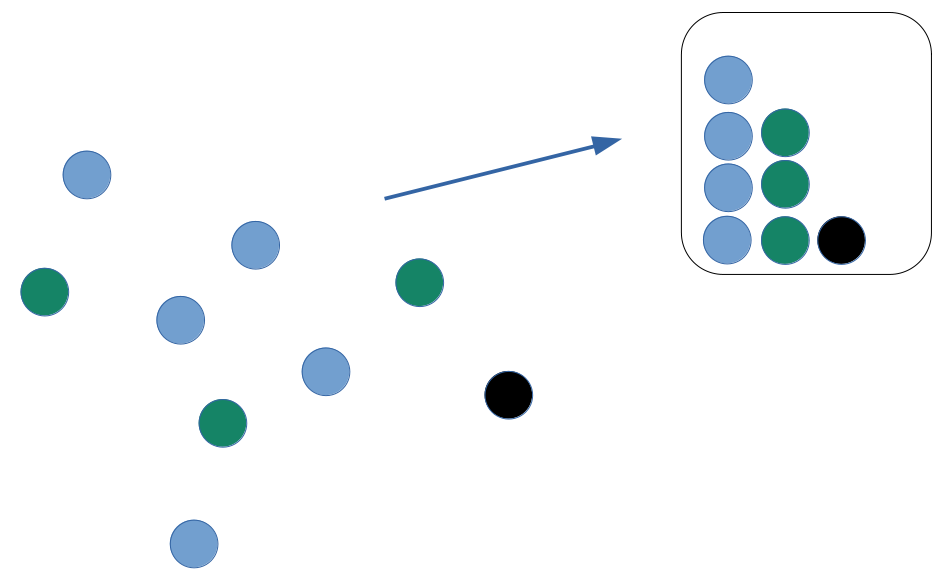
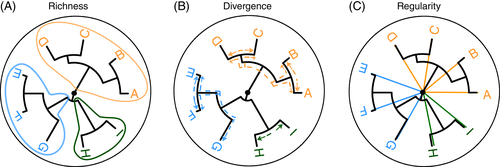
Alpha diversity indices
Richness
number of types
Eetimates of true richness based on finite sample sizes (Howard Sanders 1968); see e.g. Chao1
Evenness
- distribution of sizes (even or uneven?)
Diversity
- Combining richness & evenness
Dominance
Finite sampling
https://github.com/mblstamps/stamps2019/blob/master/STAMPS2019_overview_Pop.pdf

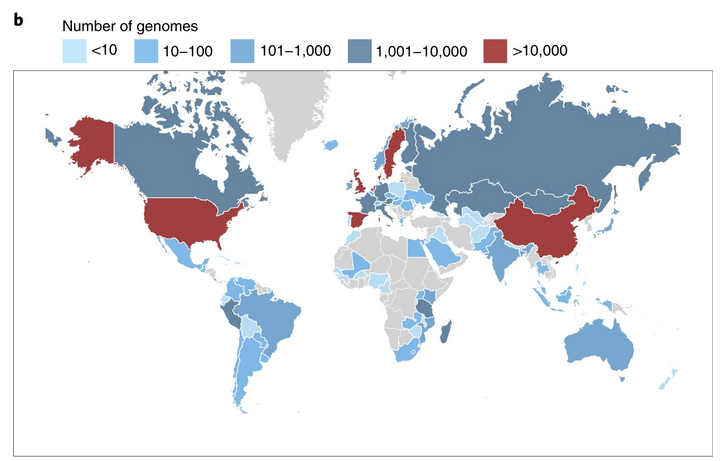
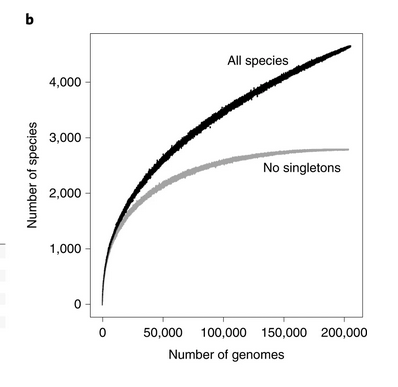
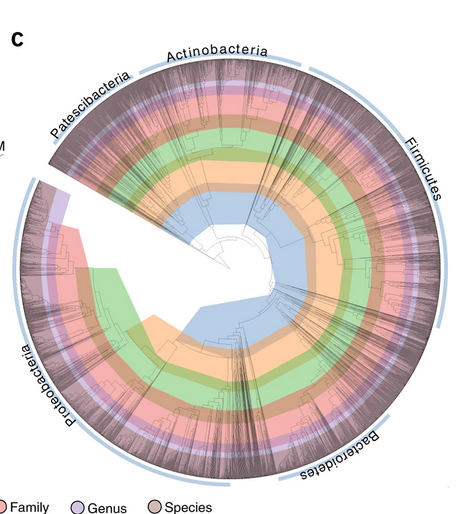
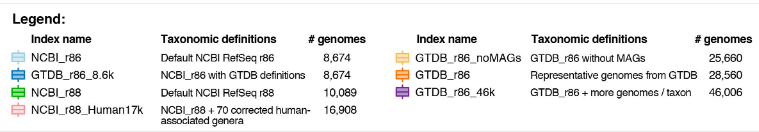
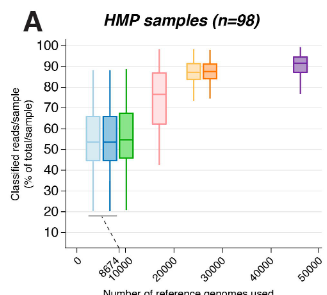
High-quality reference genomes are required for functional characterization and taxonomic assignment of the human gut microbiota.
Unified Human Gastrointestinal Genome (UHGG):
4,644 gut prokaryotes (>70% lack cultured representatives)
204,938 nonredundant genomes
Encode >170 million protein sequences, collated into Unified Human Gastrointestinal Protein (UHGP) catalog.
UHGP more than doubles the number of gut proteins in comparison to those present in the Integrated Gene Catalog.
40% of the UHGP lack functional annotations
Intraspecies genomic variation analyses revealed a large reservoir of accessory genes and single-nucleotide variants, many of which are specific to individual human populations.
The UHGG and UHGP collections enable studies linking genotypes to phenotypes in the human gut microbiome.
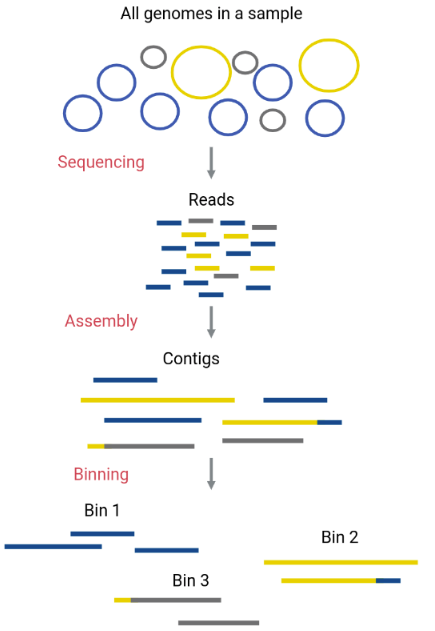
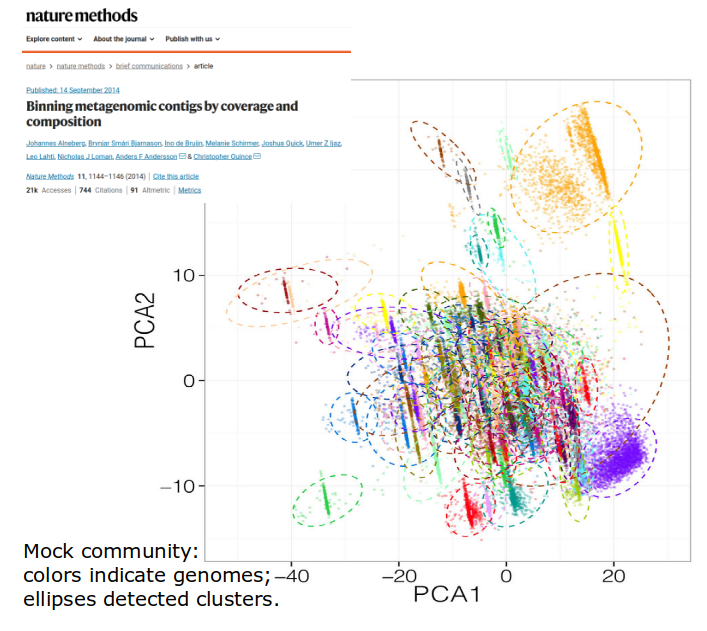
Estimating species content


Common alpha diversity indices
Phylogenetically neutral diversities:
- Richness (observed, Chao1, ACE)
- Evenness (Pielou’s evenness)
- Diversity (inverse Simpson, Shannon)
Phylogeny-aware diversities:
- Faith diversity index
Phylogenetic diversity indices
Inverse Simpson
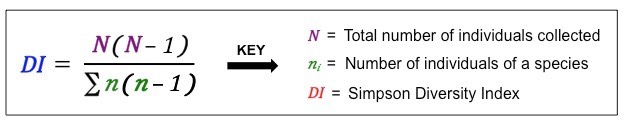
How likely it is to pick two members of the same species at random?

Inverse Simpson
Beware the variants:
Simpson (\(\lambda\))
reciprocal Simpson (\(1-\lambda\))
inverse Simpson (\(\frac{1}{\lambda}\))
Shannon diversity
Shannon Index:
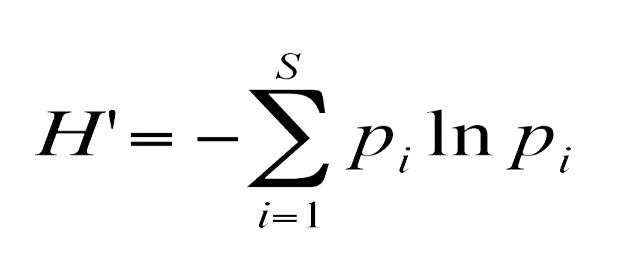
True Richness:
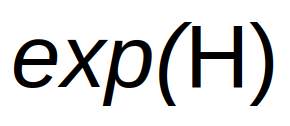
True diversity, or the effective number of types, refers to the number of equally abundant types needed for the average proportional abundance of the types to equal what is observed in the dataset of interest.
Evenness
H / ln(S)
- H: Shannon diversity
- S: Species richness
Hill’s Diversity as a unifying concept
\[\begin{equation} ^qD = (\sum_i^R p_i^q )^\frac{1}{1-q} \end{equation}\]Hill’s alpha diversities
R: richness (number of distinct types)
pi: proportion of type I
Order of diversity:
- q = 0 : Species Richness
- q = 1 : Shannon diversity
- q = 2 : (Inverse) Simpson diversity
- q ≠ 1 : Renyi entropy
Hill’s Diversity as a unifying concept
Hill’s alpha diversities
- Richness
- inverse Simpson
- Shannon
Data wrangling
Basic data operations
Transform
Subset
Merge
Aggregate
Split
Subsetting
Load example data set:
Check dimension:
Check dimension for a subset:
Transformations
- Presence/absence
- Compositional (percentages)
- \(Log_{10}\)
- CLR and other Aitchison transformations
- Phylogenetic transformations (e.g. philr)
- Custom transformations
Transformations
Task: Alternative assays
- visualize transformed data; histograms, boxplots
- compare different transformations (scatterplot?)
Agglomeration
- taxonomic units
- TreeSE objects
Agglomeration
Agglomerate microbiota data to higher taxonomic levels:
- chapter 6.3
- agglomerateByRank
- compare diversity or prevalent features between levels
Alternative experiments
Alternative assays vs. alternative experiments?
- Store agglomerated data: altExp
- Do all levels at once: splitByRanks
Splits
Splitting by:
- taxonomic units
- sample or feature groups
Taxonomic ranks & altExp
The alternative experiments (altExp) mechanism allows us to include multiple abundance tables at different taxonomic levels.
| Option | Rows (features) | Cols (samples) | Recommendation |
|---|---|---|---|
| assays | match | match | Data transformations |
| altExp | free | match | Alternative experiments |
| MultiAssay | free | free (mapping) | Multi-omic experiments |
Alternative experiments and assays?
- Pick clr assay from Genus-level data table?
- Compare Shannon diversity from Genus and Species levels?
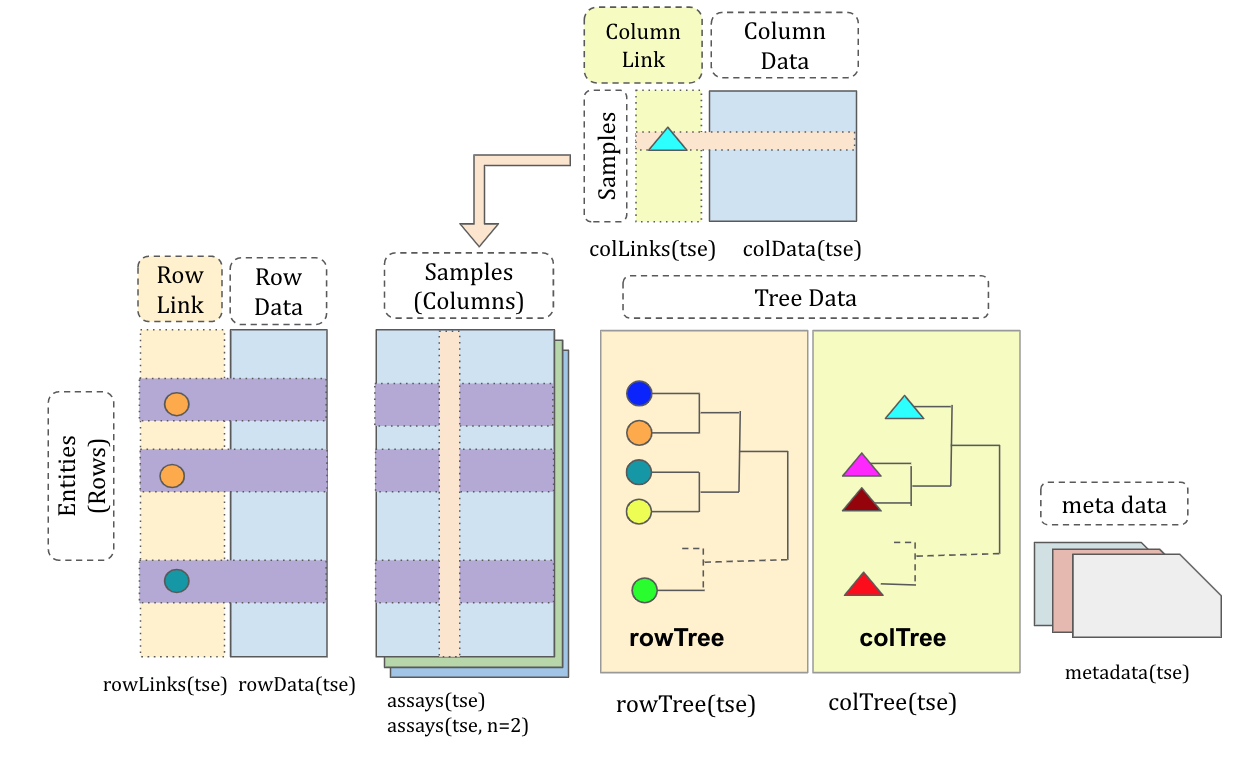
TreeSummarizedExperiment
Huang et al. F1000, 2021
Visualization
Ordination
- Visualize example data with PCoA using Bray-Curtis dissimilarity
- Visualize example data with PCA using Aitchison distance (CLR + Euclid)
Heatmaps
- Visualize abundance variation for selected taxa on a heatmap
Trees
- Visualize phylogenetic tree using the examples