These functions perform Latent Dirichlet Allocation on data stored in a
TreeSummarizedExperiment
object.
getLDA(x, ...)
addLDA(x, ...)
# S4 method for class 'SummarizedExperiment'
getLDA(x, k = 2, assay.type = "counts", eval.metric = "perplexity", ...)
# S4 method for class 'SummarizedExperiment'
addLDA(x, k = 2, assay.type = "counts", name = "LDA", ...)Arguments
- x
a
TreeSummarizedExperimentobject.- ...
optional arguments passed to
LDA- k
Integer vector. A number of latent vectors/topics. (Default:2)- assay.type
Character scalar. Specifies which assay to use for LDA ordination. (Default:"counts")- eval.metric
Character scalar. Specifies evaluation metric that will be used to select the model with the best fit. Must be either"perplexity"(topicmodels::perplexity) or"coherence"(topicdoc::topic_coherence, the best model is selected based on mean coherence). (Default:"perplexity")- name
Character scalar. The name to be used to store the result in the reducedDims of the output. (Default:"LDA")
Value
For getLDA, the ordination matrix with feature loadings matrix
as attribute "loadings".
For addLDA, a
TreeSummarizedExperiment
object is returned containing the ordination matrix in
reducedDim(..., name) with feature loadings matrix as attribute
"loadings".
Details
The functions getLDA and addLDA internally use
LDA to compute the ordination matrix and
feature loadings.
Examples
data(GlobalPatterns)
tse <- GlobalPatterns
# Reduce the number of features
tse <- agglomerateByPrevalence(tse, rank="Phylum")
# Run LDA and add the result to reducedDim(tse, "LDA")
tse <- addLDA(tse)
# Extract feature loadings
loadings <- getReducedDimAttribute(tse, "LDA", "loadings")
head(loadings)
#> 1 2
#> AD3 1.017694e-03 9.274615e-11
#> Acidobacteria 3.341907e-02 6.272831e-03
#> Actinobacteria 5.587595e-02 1.294426e-01
#> Armatimonadetes 1.825720e-04 5.268908e-04
#> BRC1 5.679833e-05 2.429880e-05
#> Bacteroidetes 2.790484e-01 9.737805e-02
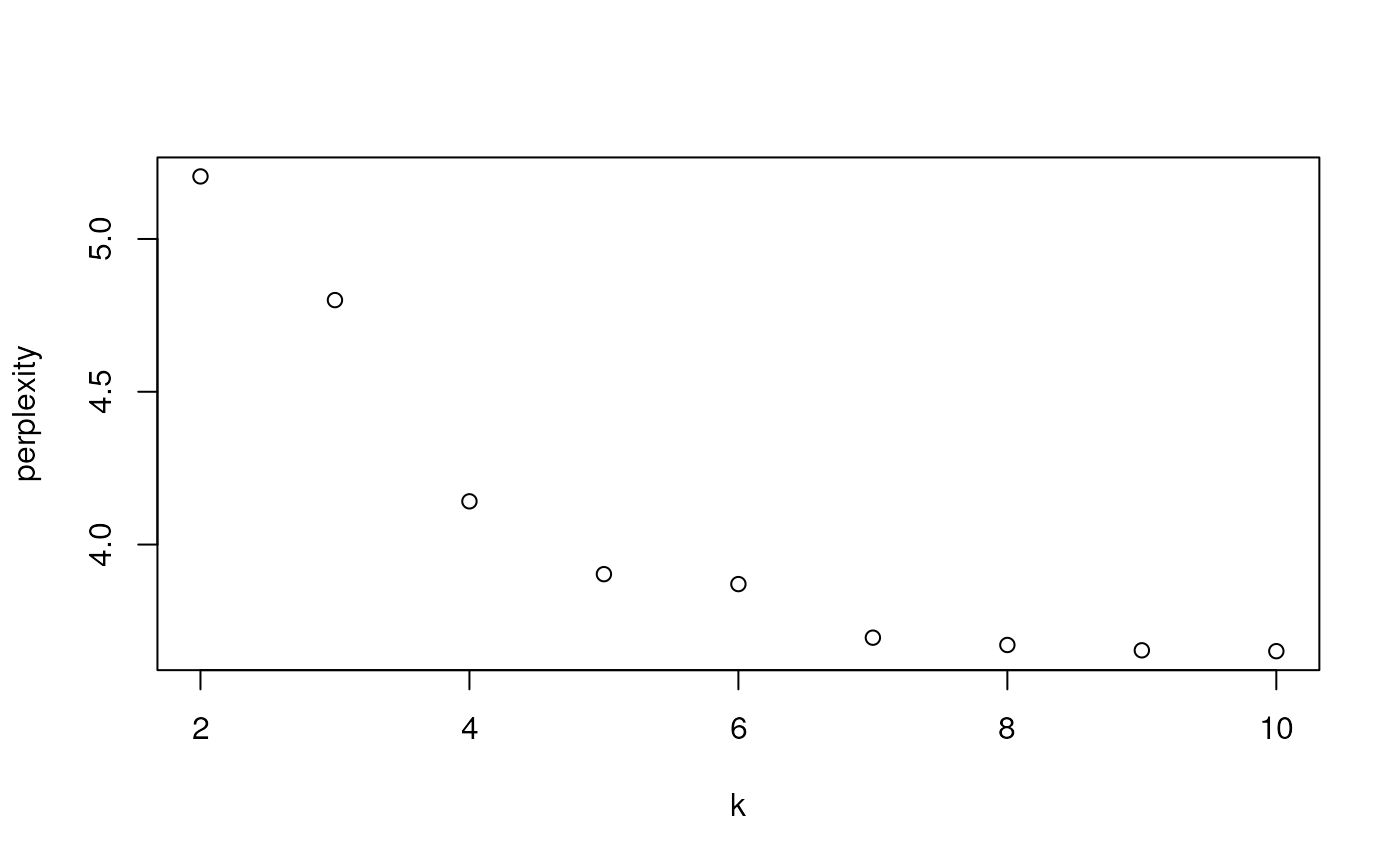
# Estimate models with number of topics from 2 to 10
tse <- addLDA(tse, k = c(2, 3, 4, 5, 6, 7, 8, 9, 10), name = "LDA_10")
# Get the evaluation metrics
tab <- getReducedDimAttribute(tse, "LDA_10","eval_metrics")
# Plot
plot(tab[["k"]], tab[["perplexity"]], xlab = "k", ylab = "perplexity")