Orchestrating Microbiome Analysis with Bioconductor (EuroBioC2024)
Source:vignettes/mia_demo.Rmd
mia_demo.RmdAuthors: Tuomas Borman1, Leo Lahti2.
Last modified:
1 September, 2024.

Overview
Description
Learn how miaverse framework integrates microbiome data science with the SummarizedExperiment ecosystem, and discover the benefits this brings. We will cover key methods, explore the OMA online book, and discuss the integration’s advantages.
We will start with a brief overview of the framework, followed by an instructor-led live demonstration. The demonstration will showcase selected key features and functionalities of miaverse. After the demo, participant will have time to discuss the framework, ask questions, and explore the tools and methods covered.
By the end of the workshop, attendees will have a understanding of how to apply the framework to their microbiome research and will be well-prepared to further explore the framework independently.
Pre-requisites
- Basic knowledge of R coding
- Familiarity with Bioconductor
- Understanding of microbiome research
If your time allows, we recommend to spend some time to explore beforehand Orchestrating Microbiome Analysis (OMA) online book.
Participation
Participants are encouraged to ask questions throughout the workshop. The session will follow a tutorial from the OMA online book, with participants running the tutorial alongside the instructor.
To facilitate this, Bioconductor provides pre-installed virtual machines with all necessary packages.
R / Bioconductor packages used
In this workshop, we will focus on the mia package, which is designed for microbiome data science. The mia package provides essential tools for analyzing microbiome data using the SummarizedExperiment framework.
Workshop goals and objectives
Learning goals
Methods for microbiome analysis: Learn to harness SummarizedExperiment ecosystem in your project.
Data structure: Understand how to utilize the TreeSummarizedExperiment class for effective microbiome data analysis.
Access resources: Get familiar with additional tools and resources, including the OMA online book.
Learning objectives
Analyze and apply methods: Apply the framework to process and analyze microbiome data.
Create visualizations: Generate and interpret visualizations.
Explore documentation: Use the OMA to explore additional tools and methods.
Workshop
1 Background
The SummarizedExperiment class is a cornerstone of Bioconductor, facilitating standardized data management across various research domains. It promotes synergy through shared methods and standardized formats, enhancing collaborative research and minimizing redundant efforts.
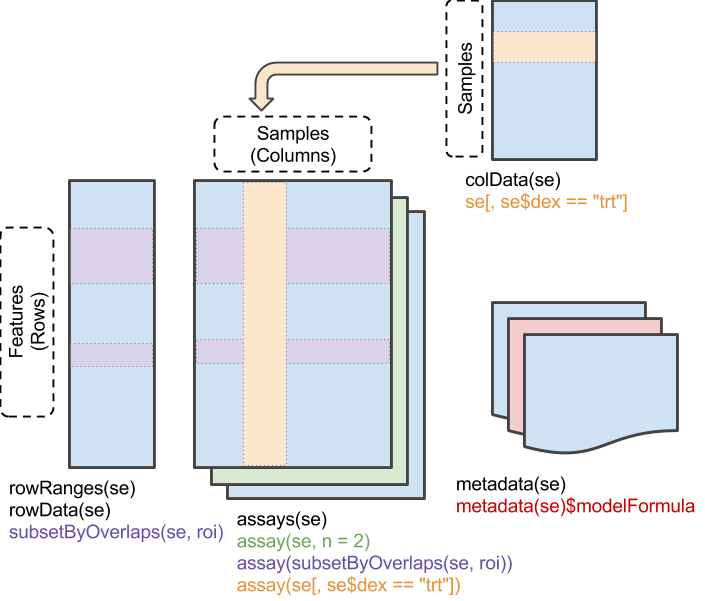
SummarizedExperiment class
To integrate microbiome data science within the SE ecosystem, the TreeSummarizedExperiment (TreeSE) was developed.
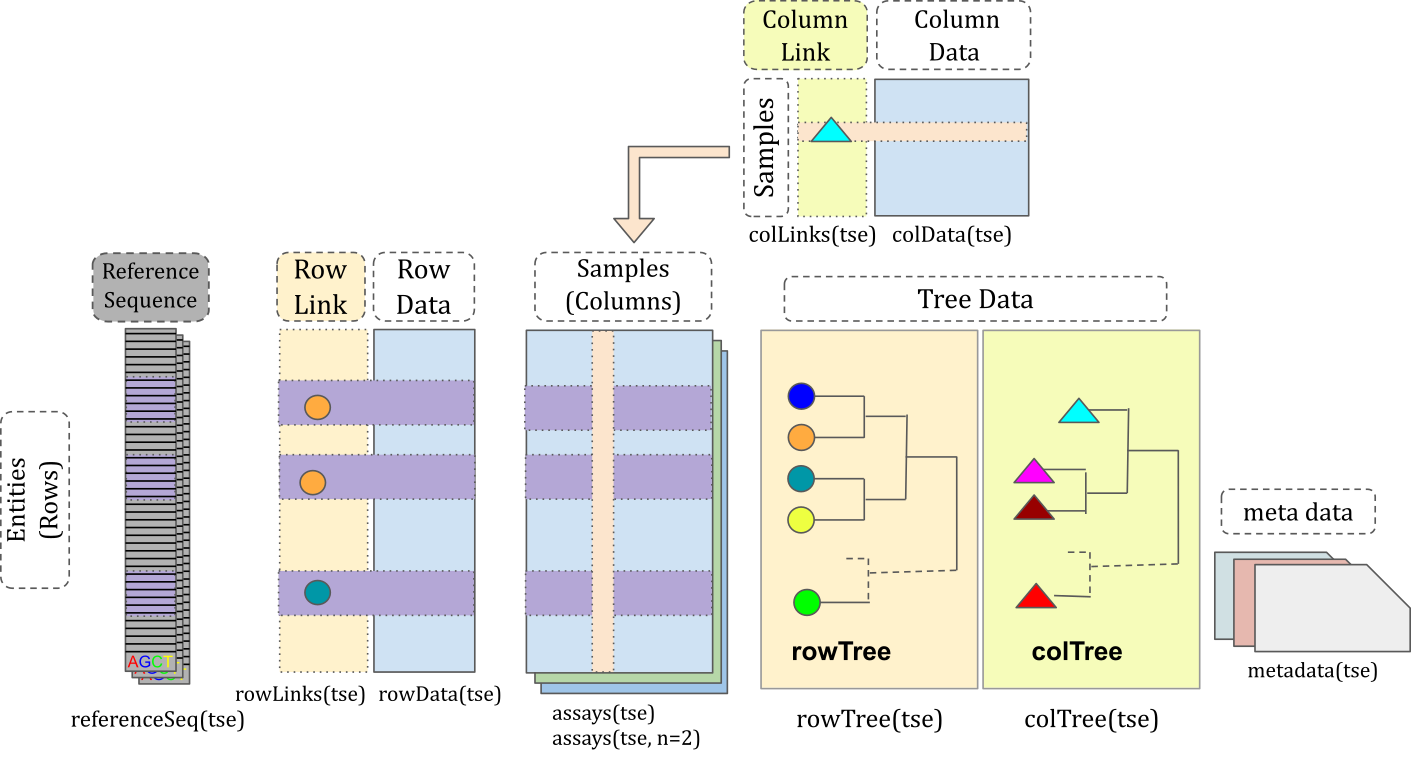
TreeSummarizedExperiment class
Subsequently, a suite of mia* (MIcrobiome Analysis) packages was developed to provide microbiome-specific methods within the SE ecosystem:
- mia: Data analysis.
- miaViz: Visualization.
- miaSim: Data simulation.
- miaTime: Time series analysis (coming soon to Bioconductor).
The framework has been further expanded by various independent developers, introducing a range of additional methods. For more details, see OMA.
To support microbiome data science, the online book Orchestrating Microbiome Analysis with Bioconductor (OMA) was created. It offers examples and best practices developed collaboratively, with plans for its upcoming publication on Bioconductor.
2 Demonstration
In this demo, we will follow the introductory workflow from OMA by copy-pasting code examples provided there. The goal is to familiarize you with TreeSE and related methods while introducing you to OMA to support continued learning beyond the workshop.
- Set up your local computer or access an online environment.
- Go to the introductory workflow.
- Be ready to follow along with the demonstration.
3 Q&A and free exploration
Time for discussion. You are also encouraged to explore and run examples from OMA on your own.
Thank you for your time!
Thank you for participating in the workshop! Stay tuned for the upcoming publication of miaverse and continue exploring its capabilities.
For any future questions or additional support, please refer to the OMA support page.
University of Turku, tvborm@utu.fi↩︎
University of Turku↩︎