Data containers in R/Bioconductor
Stuctured way to represent data
- Biological data cannot be represented with a single table
- Managing multiple tables becomes easily a bottleneck of efficient workflows


Standardized data containers
Central for the R/Bioconductor ecosystem: phyloseq, (Tree)SummarizedExperiment, MultiAssayExperiment

Data containers support collaborative workflow development

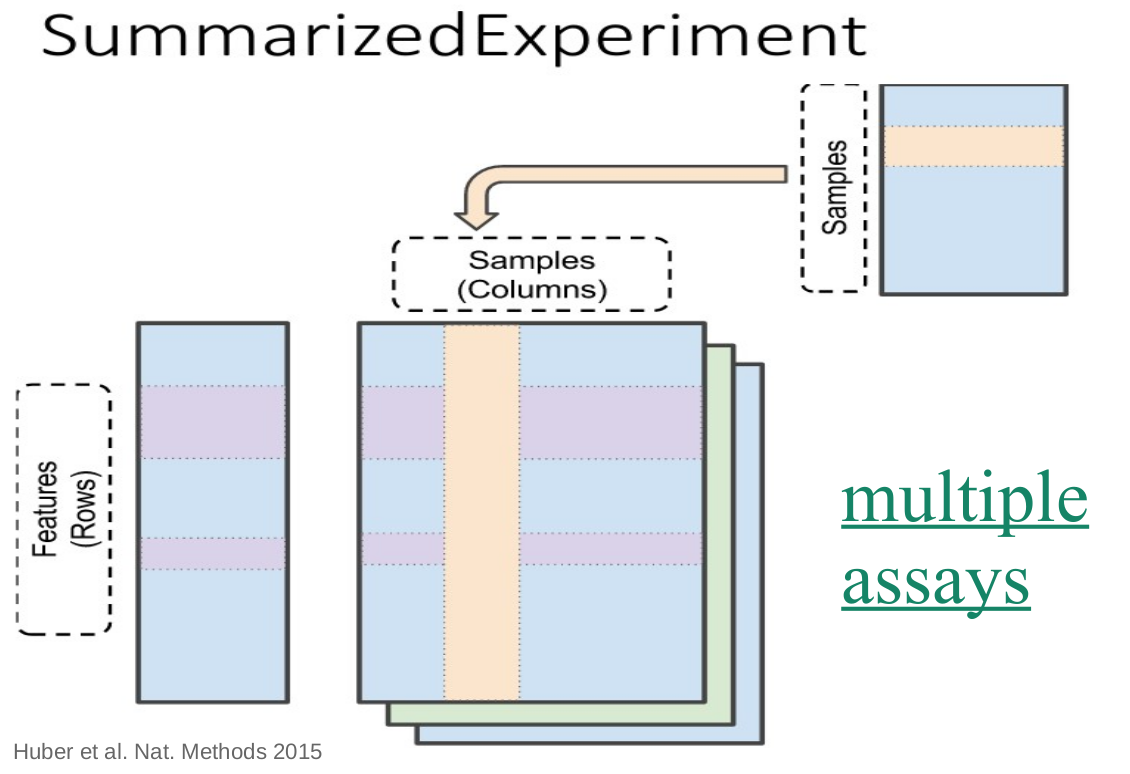
SummarizedExperiment
- Most common data container
- Optimized for biological data
- Extended to different purposes




Optimal container for microbiome data?
Optimal container for microbiome data?
- Multiple assays: seamless interlinking
Optimal container for microbiome data?
- Multiple assays: seamless interlinking
- Hierarchical data: supporting samples & features
Optimal container for microbiome data?
- Multiple assays: seamless interlinking
- Hierarchical data: supporting samples & features
- Side information: extended capabilities & data types
Optimal container for microbiome data?
- Multiple assays: seamless interlinking
- Hierarchical data: supporting samples & features
- Side information: extended capabilities & data types
- Optimized: for speed & memory
Optimal container for microbiome data?
- Multiple assays: seamless interlinking
- Hierarchical data: supporting samples & features
- Side information: extended capabilities & data types
- Optimized: for speed & memory
- Integrated: with other applications & frameworks
Optimal container for microbiome data?
- Multiple assays: seamless interlinking
- Hierarchical data: supporting samples & features
- Side information: extended capabilities & data types
- Optimized: for speed & memory
- Integrated: with other applications & frameworks
Reduce overlapping efforts, improve interoperability, ensure sustainability.
phyloseq
- The first microbome data container from around 2010.
- Has become standard for (16S) microbiome bioinformatics in R (J McMurdie, S Holmes et al.)

TreeSummarizedExperiment
New, alternative microbiome data container.
- Extension to SummarizedExperiment
- Optimal for microbiome data
- Links microbiome field to larger SummarizedExperiment family
Huang et al. F1000, 2021


Orchestrating Microbiome Analysis with R and Bioconductor – online book: beta version
Current framework
- (Tree)SummarizedExperiment for single omics
- MultiAssayExperiment for multi-omics
MultiAssayExperiment
- Links (Tree)SummarizedExperiment objects

Ramos et al. Cancer Res., 2017
Task: load microbiome data
Load an example data set from the mia R package with:
Source: Hintikka et al. (2021). Xylo-oligosaccharides in prevention of hepatic steatosis and adipose tissue inflammation: Associating taxonomic and metabolomic patterns in fecal microbiomes with biclustering. International Journal of Environmental Research and Public Health 18(8) https://doi.org/10.3390/ijerph18084049
Task: load microbiome data
This is MultiAssayExperiment data object. Let us check what experiment it contains.
Task: load microbiome data
Let us pick the microbiota data, which is TreeSummarizedExperiment object.
class: TreeSummarizedExperiment
dim: 12706 40
metadata(0):
assays(1): counts
rownames(12706): GAYR01026362.62.2014 CVJT01000011.50.2173 ...
JRJTB:03787:02429 JRJTB:03787:02478
rowData names(7): Phylum Class ... Species OTU
colnames(40): C1 C2 ... C39 C40
colData names(0):
reducedDimNames(0):
mainExpName: NULL
altExpNames(0):
rowLinks: NULL
rowTree: NULL
colLinks: NULL
colTree: NULLOpen microbiome data resources
Open microbiome data resources supporting TreeSummarizedExperiment:
- R package data (mia, miaViz, miaTime)
- Human studies: curatedMetagenomicData (Pasolli et al. Nat Meth 2017)
- EBI MGnify: MGnifyR R package
- Other studies: microbiomeDataSets (Lahti et al.)
See also OMA chapter on available data sets.