17 Microbe Set Enrichment Analysis (MSEA)
Similar to gene set enrichment analyses for genes (Subramanian et al. 2005), an obvious next step following differential abundance analysis in microbiome studies is to conduct enrichment analysis for microbe sets, known as microbe set enrichment analysis (MSEA) (Kou et al. 2020). Similar to GSEA, the primary goal of MSEA is to detect the modest but coordinated changes in pre-specified sets of related microbial features. Such a set might include all the microbes in a specific pathway or microbial genes that have been shown to be co-regulated based on previously published studies. Like GSEA, MSEA aggregates the per-feature statistics across microbes within a microbe set. This corresponds to the hypothesis that many relevant phenotype differences are manifested by small but consistent changes in a set of features.
The goal of the MSEA approach is to determine if the members of S (microbe set) are randomly distributed throughout the ranked list of features (L) or primarily found at the top or bottom. We will use the R package gsEasy to conduct the MSEA test described by Subramanian et al. (2005).
17.1 Input data for MSEA using species relative abundance data
In this chapter, we will use the publicly available Inflammatory Bowel Diseases (IBD) microbiome data from the integrative Human Microbiome Project (iHMP) available from the curatedMetagenomicData package (Lloyd-Price et al. 2019). We aim to conduct MSEA analysis based on both taxonomic profiles (species relative abundances) and functional profiles (pathway relative abundances).
17.2 Performing the MSEA analysis with species relative abundance data
We will first prepare the input feature table and sample metadata for differential abundance analysis using MaAsLin2 (Mallick et al. 2021). The ranked feature list from the differential abundance analysis serves as an input for the MSEA.
##################
# Load iHMP data #
##################
library(curatedMetagenomicData)
library(dplyr)
se_relative <- sampleMetadata |>
filter(study_name == "HMP_2019_ibdmdb") |>
returnSamples("relative_abundance", rownames = "short")
##########################
# Create sample metadata #
##########################
sample_metadata <-
colData(se_relative) %>%
as.data.frame() %>% filter(visit_number == 1) %>%
.[, c("age", "disease", "antibiotics_current_use")]
#################
# Set reference #
#################
sample_metadata$disease <- as.factor(sample_metadata$disease)
sample_metadata$disease <- relevel(sample_metadata$disease, 'healthy')
###########################
# Create species features #
###########################
feature_species_t <- as.data.frame(assay(se_relative))
rownames(feature_species_t) <- sub('.*s__', '', rownames(feature_species_t))
##############################
# Subset to baseline samples #
##############################
feature_species <- as.data.frame(t(feature_species_t))
feature_species <- feature_species[rownames(sample_metadata),]
feature_species <- feature_species / 100
rm(feature_species_t); rm(se_relative)In the next step, we will use MaAsLin2 to fit a multivariable regression model for testing the association between microbial species abundance versus IBD diagnosis. The analysis method we use here is “LM”, which is the default setting. We also adjust for age and antibiotic usage, following the original study.
library(Maaslin2)
fit_data = Maaslin2(input_data = feature_species,
input_metadata = sample_metadata,
normalization = "NONE",
output = "output_species",
fixed_effects = c("disease", "age", "antibiotics_current_use"))
## [1] "Creating output folder"
## [1] "Creating output feature tables folder"
## [1] "Creating output fits folder"
## [1] "Creating output figures folder"
## 2024-07-16 13:50:59.818804 INFO::Writing function arguments to log file
## 2024-07-16 13:50:59.833449 INFO::Verifying options selected are valid
## 2024-07-16 13:50:59.860259 INFO::Determining format of input files
## 2024-07-16 13:50:59.860768 INFO::Input format is data samples as rows and metadata samples as rows
## 2024-07-16 13:50:59.866296 INFO::Formula for fixed effects: expr ~ disease + age + antibiotics_current_use
## 2024-07-16 13:50:59.866958 INFO::Factor detected for categorial metadata 'disease'. Provide a reference argument or manually set factor ordering to change reference level.
## 2024-07-16 13:50:59.867582 INFO::Filter data based on min abundance and min prevalence
## 2024-07-16 13:50:59.868023 INFO::Total samples in data: 136
## 2024-07-16 13:50:59.868465 INFO::Min samples required with min abundance for a feature not to be filtered: 13.600000
## 2024-07-16 13:50:59.874153 INFO::Total filtered features: 452
## 2024-07-16 13:50:59.874832 INFO::Filtered feature names from abundance and prevalence filtering: Abiotrophia.sp..HMSC24B09, Acidaminococcus.sp..CAG.542, Acinetobacter.lwoffii, Acinetobacter.ursingii, Actinobaculum.sp..oral.taxon.183, Actinomyces.graevenitzii, Actinomyces.sp..HMSC035G02, Actinomyces.sp..HPA0247, Actinomyces.sp..ICM47, Actinomyces.sp..oral.taxon.180, Actinomyces.sp..oral.taxon.181, Actinomyces.urogenitalis, Actinotignum.timonense, Adlercreutzia.caecimuris, Adlercreutzia.equolifaciens, Aeriscardovia.aeriphila, Aerococcus.urinaeequi, Aeromonas.dhakensis, Aeromonas.hydrophila, Aggregatibacter.aphrophilus, Aggregatibacter.segnis, Aggregatibacter.sp..oral.taxon.458, Alistipes.inops, Alistipes.onderdonkii, Alistipes.timonensis, Allisonella.histaminiformans, Alloprevotella.tannerae, Alloscardovia.omnicolens, Amedibacillus.dolichus, Anaerococcus.lactolyticus, Anaerococcus.vaginalis, Anaerocolumna.aminovalerica, Anaerofilum.sp..An201, Anaerofustis.stercorihominis, Anaeroglobus.geminatus, Anaeromassilibacillus.sp..An172, Anaerosporobacter.mobilis, Anaerostipes.caccae, Anaerostipes.sp..494a, Anaerostipes.sp..992a, Anaerotruncus.sp..CAG.528, Arthrospira.platensis, Atopobium.deltae, Atopobium.minutum, Bacteroidales.bacterium.KA00251, Bacteroides.clarus, Bacteroides.fluxus, Bacteroides.nordii, Bacteroides.oleiciplenus, Bacteroides.sp..43_108, Bacteroides.sp..CAG.144, Bacteroides.sp..CAG.530, Bacteroides.sp..CAG.598, Bacteroides.sp..CAG.633, Bacteroides.sp..CAG.661, Bacteroides.sp..D2, Bacteroides.sp..OM08.11, Bacteroides.stercorirosoris, Bacteroidetes.oral.taxon.274, Bavariicoccus.seileri, Bifidobacterium.angulatum, Bifidobacterium.animalis, Bifidobacterium.asteroides, Bifidobacterium.breve, Bifidobacterium.catenulatum, Bifidobacterium.dentium, Bifidobacterium.pseudolongum, Bifidobacterium.pullorum, Blastocystis.sp..subtype.1, Blautia.argi, Blautia.coccoides, Blautia.hansenii, Blautia.hydrogenotrophica, Blautia.producta, Brachyspira.pilosicoli, Butyricicoccus.pullicaecorum, Butyrivibrio.crossotus, Butyrivibrio.sp..CAG.318, Campylobacter.concisus, Campylobacter.gracilis, Campylobacter.hominis, Campylobacter.showae, Campylobacter.upsaliensis, Campylobacter.ureolyticus, Candidatus.Gastranaerophilales.bacterium, Candidatus.Methanomassiliicoccus.intestinalis, Candidatus.Stoquefichus.sp..KLE1796, Catenibacterium.mitsuokai, Cellulosilyticum.lentocellum, Chlamydia.ibidis, Christensenella.hongkongensis, Christensenella.minuta, Citrobacter.amalonaticus, Citrobacter.braakii, Citrobacter.europaeus, Citrobacter.farmeri, Citrobacter.freundii, Citrobacter.pasteurii, Citrobacter.portucalensis, Citrobacter.werkmanii, Citrobacter.youngae, Cloacibacillus.porcorum, Clostridiales.bacterium.1_7_47FAA, Clostridiales.bacterium.CHKCI006, Clostridioides.difficile, Clostridium.baratii, Clostridium.botulinum, Clostridium.butyricum, Clostridium.cadaveris, Clostridium.celatum, Clostridium.disporicum, Clostridium.neonatale, Clostridium.paraputrificum, Clostridium.perfringens, Clostridium.sp..7_2_43FAA, Clostridium.sp..CAG.167, Clostridium.sp..CAG.242, Clostridium.sp..CAG.253, Clostridium.sp..CAG.411, Clostridium.sp..CAG.413, Clostridium.sp..CAG.590, Clostridium.sp..CAG.678, Clostridium.sp..CAG.964, Clostridium.sp..D5, Clostridium.sp..MSTE9, Clostridium.sp..chh4.2, Clostridium.sporogenes, Comamonas.kerstersii, Coprobacillus.cateniformis, Coprobacter.secundus, Coprobacter.sp., Corynebacterium.accolens, Corynebacterium.amycolatum, Corynebacterium.kroppenstedtii, Corynebacterium.matruchotii, Corynebacterium.oculi, Cronobacter.malonaticus, Cronobacter.sakazakii, Cutibacterium.acnes, Cutibacterium.granulosum, Dellaglioa.algida, Desulfovibrio.fairfieldensis, Desulfovibrio.piger, Desulfovibrionaceae.bacterium, Dialister.micraerophilus, Dialister.pneumosintes, Dialister.sp..CAG.357, Dorea.sp..CAG.317, Dysgonomonas.gadei, Dysgonomonas.mossii, Dysgonomonas.sp..37.18, Eikenella.corrodens, Enhydrobacter.aerosaccus, Enorma.massiliensis, Enterobacter.mori, Enterocloster.asparagiformis, Enterococcus.asini, Enterococcus.avium, Enterococcus.casseliflavus, Enterococcus.dispar, Enterococcus.durans, Enterococcus.faecalis, Enterococcus.faecium, Enterococcus.gallinarum, Enterococcus.hirae, Enterococcus.mundtii, Enterococcus.pseudoavium, Enterococcus.raffinosus, Enterococcus.sp..3H8_DIV0648, Enterococcus.thailandicus, Erysipelothrix.larvae, Escherichia.albertii, Escherichia.marmotae, Eubacteriaceae.bacterium.CHKCI005, Eubacterium.coprostanoligenes, Eubacterium.dolichum.CAG.375, Eubacterium.limosum, Eubacterium.sp..An11, Eubacterium.sp..CAG.180, Eubacterium.sp..CAG.274, Eubacterium.sp..OM08.24, Ezakiella.coagulans, Faecalicatena.orotica, Faecalicoccus.pleomorphus, Faecalitalea.cylindroides, Fannyhessea.vaginae, Finegoldia.magna, Firmicutes.bacterium.CAG.110, Firmicutes.bacterium.CAG.145, Firmicutes.bacterium.CAG.170, Firmicutes.bacterium.CAG.238, Firmicutes.bacterium.CAG.424, Firmicutes.bacterium.CAG.534, Firmicutes.bacterium.CAG.646, Firmicutes.bacterium.CAG.95, Flavonifractor.sp..An10, Flavonifractor.sp..An100, Flavonifractor.sp..An306, Fretibacterium.fastidiosum, Frigoribacterium.sp..Leaf8, Fructilactobacillus.sanfranciscensis, Fusobacterium.equinum, Fusobacterium.gonidiaformans, Fusobacterium.mortiferum, Fusobacterium.naviforme, Fusobacterium.nucleatum, Fusobacterium.periodonticum, Fusobacterium.sp..CAG.439, Fusobacterium.sp..oral.taxon.370, Fusobacterium.ulcerans, Gemella.asaccharolytica, Gemella.haemolysans, Gemella.morbillorum, Gemella.sanguinis, Gemmiger.sp..An50, Gemmiger.sp..An87, Gleimia.europaea, Gordonibacter.pamelaeae, Granulicatella.adiacens, Haemophilus.haemolyticus, Haemophilus.influenzae, Haemophilus.parahaemolyticus, Haemophilus.paraphrohaemolyticus, Haemophilus.pittmaniae, Haemophilus.sputorum, Hafnia.alvei, Hafnia.paralvei, Harryflintia.acetispora, Holdemanella.biformis, Klebsiella.aerogenes, Klebsiella.michiganensis, Klebsiella.oxytoca, Klebsiella.quasipneumoniae, Klebsiella.variicola.CAG.634, Kluyvera.ascorbata, Kluyvera.cryocrescens, Kluyvera.georgiana, Kocuria.palustris, Kosakonia.sacchari, Lachnoclostridium.sp..An118, Lachnoclostridium.sp..An131, Lachnoclostridium.sp..An138, Lachnoclostridium.sp..An14, Lachnoclostridium.sp..An181, Lachnoclostridium.sp..An298, Lachnospiraceae.bacterium.2_1_46FAA, Lachnospiraceae.bacterium.oral.taxon.096, Lacrimispora.celerecrescens, Lacticaseibacillus.rhamnosus, Lactiplantibacillus.plantarum, Lactobacillus.acidophilus, Lactobacillus.amylovorus, Lactobacillus.crispatus, Lactobacillus.delbrueckii, Lactobacillus.gasseri, Lactobacillus.jensenii, Lactobacillus.johnsonii, Lactobacillus.paragasseri, Lactococcus.lactis, Lactococcus.petauri, Lactococcus.piscium, Lactonifactor.longoviformis, Lancefieldella.parvula, Lancefieldella.rimae, Latilactobacillus.sakei, Lawsonella.clevelandensis, Leclercia.adecarboxylata, Leuconostoc.garlicum, Leuconostoc.lactis, Ligilactobacillus.animalis, Ligilactobacillus.ruminis, Ligilactobacillus.salivarius, Limosilactobacillus.fermentum, Limosilactobacillus.mucosae, Limosilactobacillus.oris, Limosilactobacillus.reuteri, Limosilactobacillus.vaginalis, Massilimicrobiota.timonensis, Megamonas.funiformis, Megamonas.funiformis.CAG.377, Megamonas.hypermegale, Megasphaera.elsdenii, Megasphaera.micronuciformis, Megasphaera.sp..DISK.18, Megasphaera.sp..MJR8396C, Methanobrevibacter.smithii, Micrococcus.aloeverae, Micrococcus.luteus, Micrococcus.lylae, Microvirgula.aerodenitrificans, Mitsuokella.jalaludinii, Mitsuokella.multacida, Moraxella.osloensis, Morganella.morganii, Murimonas.intestini, Neisseria.cinerea, Neisseria.flavescens, Neisseria.sp..oral.taxon.014, Neisseria.subflava, Obesumbacterium.proteus, Odoribacter.laneus, Oscillibacter.sp..PC13, Oxalobacter.formigenes, Paenibacillus.macerans, Parabacteroides.goldsteinii, Parabacteroides.johnsonii, Parabacteroides.sp..CAG.409, Pararheinheimera.mesophila, Parvimonas.micra, Parvimonas.sp..KA00067, Parvimonas.sp..oral.taxon.110, Parvimonas.sp..oral.taxon.393, Paucilactobacillus.vaccinostercus, Pediococcus.acidilactici, Pediococcus.pentosaceus, Peptococcus.niger, Peptoniphilus.coxii, Peptoniphilus.duerdenii, Peptoniphilus.harei, Peptoniphilus.lacrimalis, Peptoniphilus.sp..BV3C26, Peptoniphilus.sp..HMSC062D09, Peptostreptococcus.anaerobius, Peptostreptococcus.stomatis, Phascolarctobacterium.sp..CAG.266, Phascolarctobacterium.succinatutens, Phocaeicola.sartorii, Phytobacter.palmae, Porphyromonas.asaccharolytica, Porphyromonas.endodontalis, Porphyromonas.somerae, Porphyromonas.sp..HMSC065F10, Porphyromonas.uenonis, Prevotella.amnii, Prevotella.bergensis, Prevotella.bivia, Prevotella.buccae, Prevotella.buccalis, Prevotella.colorans, Prevotella.corporis, Prevotella.dentalis, Prevotella.denticola, Prevotella.disiens, Prevotella.histicola, Prevotella.intermedia, Prevotella.jejuni, Prevotella.melaninogenica, Prevotella.nigrescens, Prevotella.oralis, Prevotella.oris, Prevotella.oulorum, Prevotella.pallens, Prevotella.salivae, Prevotella.sp..885, Prevotella.sp..AM42.24, Prevotella.sp..CAG.1092, Prevotella.sp..CAG.1185, Prevotella.sp..CAG.279, Prevotella.sp..CAG.520, Prevotella.sp..CAG.891, Prevotella.sp..S7.1.8, Prevotella.stercorea, Prevotella.timonensis, Proteus.hauseri, Proteus.mirabilis, Proteus.penneri, Proteus.vulgaris, Providencia.alcalifaciens, Pseudoflavonifractor.capillosus, Pseudoflavonifractor.sp..An184, Pseudoflavonifractor.sp..An85, Pseudomonas.fragi, Pseudomonas.guguanensis, Raoultella.ornithinolytica, Raoultella.planticola, Rikenella.microfusus, Robinsoniella.sp..RHS, Romboutsia.ilealis, Roseburia.sp..CAG.182, Roseburia.sp..CAG.303, Roseburia.sp..CAG.309, Rothia.mucilaginosa, Ruminococcaceae.bacterium.D5, Ruminococcus.callidus, Ruminococcus.obeum.CAG.39, Ruminococcus.sp..CAG.330, Ruminococcus.sp..CAG.403, Ruminococcus.sp..CAG.488, Ruminococcus.sp..CAG.563, Saccharomyces.cerevisiae, Sanguibacteroides.justesenii, Sarcina.ventriculi, Scardovia.wiggsiae, Schaalia.odontolytica, Schaalia.turicensis, Serratia.liquefaciens, Serratia.marcescens, Sharpea.azabuensis, Shuttleworthia.satelles, Slackia.isoflavoniconvertens, Solobacterium.moorei, Staphylococcus.aureus, Staphylococcus.epidermidis, Staphylococcus.hominis, Streptococcus.agalactiae, Streptococcus.australis, Streptococcus.cristatus, Streptococcus.gallolyticus, Streptococcus.gordonii, Streptococcus.infantis, Streptococcus.lutetiensis, Streptococcus.macedonicus, Streptococcus.milleri, Streptococcus.mitis, Streptococcus.mutans, Streptococcus.oralis, Streptococcus.pasteurianus, Streptococcus.peroris, Streptococcus.salivarius.CAG.79, Streptococcus.sanguinis, Streptococcus.sp..A12, Streptococcus.sp..F0442, Streptococcus.sp..HPH0090, Streptococcus.sp..M334, Streptococcus.thermophilus, Streptococcus.vestibularis, Terrisporobacter.othiniensis, Thermoleophilum.album, Tractidigestivibacter.scatoligenes, Treponema.lecithinolyticum, Turicibacter.sanguinis, Tyzzerella.nexilis, Varibaculum.cambriense, Veillonella.rodentium, Veillonella.rogosae, Veillonella.seminalis, Veillonella.sp..CAG.933, Veillonella.tobetsuensis, Victivallis.vadensis, Weissella.cibaria, Weissella.confusa, Weissella.viridescens, Wohlfahrtiimonas.chitiniclastica, Yersinia.frederiksenii, X.Bacteroides..pectinophilus, X.Butyribacterium..methylotrophicum, X.Clostridium..hylemonae, X.Clostridium..methylpentosum, X.Clostridium..scindens, X.Clostridium..spiroforme, X.Eubacterium..brachy, X.Eubacterium..infirmum
## 2024-07-16 13:50:59.877519 INFO::Total filtered features with variance filtering: 0
## 2024-07-16 13:50:59.878001 INFO::Filtered feature names from variance filtering:
## 2024-07-16 13:50:59.878419 INFO::Running selected normalization method: NONE
## 2024-07-16 13:50:59.878886 INFO::Applying z-score to standardize continuous metadata
## 2024-07-16 13:50:59.883009 INFO::Running selected transform method: LOG
## 2024-07-16 13:50:59.885849 INFO::Running selected analysis method: LM
## 2024-07-16 13:50:59.889283 INFO::Fitting model to feature number 1, Acidaminococcus.intestini
## 2024-07-16 13:50:59.895365 INFO::Fitting model to feature number 2, Agathobaculum.butyriciproducens
## 2024-07-16 13:50:59.897751 INFO::Fitting model to feature number 3, Akkermansia.muciniphila
## 2024-07-16 13:50:59.900096 INFO::Fitting model to feature number 4, Alistipes.finegoldii
## 2024-07-16 13:50:59.902342 INFO::Fitting model to feature number 5, Alistipes.indistinctus
## 2024-07-16 13:50:59.904621 INFO::Fitting model to feature number 6, Alistipes.putredinis
## 2024-07-16 13:50:59.906837 INFO::Fitting model to feature number 7, Alistipes.shahii
## 2024-07-16 13:50:59.909104 INFO::Fitting model to feature number 8, Anaerobutyricum.hallii
## 2024-07-16 13:50:59.911315 INFO::Fitting model to feature number 9, Anaeromassilibacillus.sp..An250
## 2024-07-16 13:50:59.913628 INFO::Fitting model to feature number 10, Anaerostipes.hadrus
## 2024-07-16 13:50:59.915891 INFO::Fitting model to feature number 11, Anaerotignum.lactatifermentans
## 2024-07-16 13:50:59.918116 INFO::Fitting model to feature number 12, Anaerotruncus.colihominis
## 2024-07-16 13:50:59.920388 INFO::Fitting model to feature number 13, Bacteroides.caccae
## 2024-07-16 13:50:59.922613 INFO::Fitting model to feature number 14, Bacteroides.cellulosilyticus
## 2024-07-16 13:50:59.924862 INFO::Fitting model to feature number 15, Bacteroides.eggerthii
## 2024-07-16 13:50:59.927084 INFO::Fitting model to feature number 16, Bacteroides.faecis
## 2024-07-16 13:50:59.929351 INFO::Fitting model to feature number 17, Bacteroides.faecis.CAG.32
## 2024-07-16 13:50:59.931621 INFO::Fitting model to feature number 18, Bacteroides.finegoldii
## 2024-07-16 13:50:59.933835 INFO::Fitting model to feature number 19, Bacteroides.fragilis
## 2024-07-16 13:50:59.936115 INFO::Fitting model to feature number 20, Bacteroides.galacturonicus
## 2024-07-16 13:50:59.938327 INFO::Fitting model to feature number 21, Bacteroides.intestinalis
## 2024-07-16 13:50:59.940569 INFO::Fitting model to feature number 22, Bacteroides.ovatus
## 2024-07-16 13:50:59.94278 INFO::Fitting model to feature number 23, Bacteroides.salyersiae
## 2024-07-16 13:50:59.945058 INFO::Fitting model to feature number 24, Bacteroides.stercoris
## 2024-07-16 13:50:59.947336 INFO::Fitting model to feature number 25, Bacteroides.thetaiotaomicron
## 2024-07-16 13:50:59.949614 INFO::Fitting model to feature number 26, Bacteroides.uniformis
## 2024-07-16 13:50:59.951884 INFO::Fitting model to feature number 27, Bacteroides.xylanisolvens
## 2024-07-16 13:50:59.954106 INFO::Fitting model to feature number 28, Barnesiella.intestinihominis
## 2024-07-16 13:50:59.956372 INFO::Fitting model to feature number 29, Bifidobacterium.adolescentis
## 2024-07-16 13:50:59.958607 INFO::Fitting model to feature number 30, Bifidobacterium.bifidum
## 2024-07-16 13:50:59.96088 INFO::Fitting model to feature number 31, Bifidobacterium.longum
## 2024-07-16 13:50:59.963102 INFO::Fitting model to feature number 32, Bifidobacterium.pseudocatenulatum
## 2024-07-16 13:50:59.965334 INFO::Fitting model to feature number 33, Bilophila.wadsworthia
## 2024-07-16 13:50:59.967707 INFO::Fitting model to feature number 34, Blautia.obeum
## 2024-07-16 13:50:59.96995 INFO::Fitting model to feature number 35, Blautia.sp..CAG.257
## 2024-07-16 13:50:59.972285 INFO::Fitting model to feature number 36, Blautia.wexlerae
## 2024-07-16 13:50:59.97451 INFO::Fitting model to feature number 37, Butyricimonas.synergistica
## 2024-07-16 13:50:59.976806 INFO::Fitting model to feature number 38, Butyricimonas.virosa
## 2024-07-16 13:50:59.979068 INFO::Fitting model to feature number 39, Clostridium.bolteae.CAG.59
## 2024-07-16 13:50:59.981317 INFO::Fitting model to feature number 40, Clostridium.sp..CAG.299
## 2024-07-16 13:50:59.983596 INFO::Fitting model to feature number 41, Clostridium.sp..CAG.58
## 2024-07-16 13:50:59.98586 INFO::Fitting model to feature number 42, Collinsella.aerofaciens
## 2024-07-16 13:50:59.988123 INFO::Fitting model to feature number 43, Collinsella.intestinalis
## 2024-07-16 13:50:59.990353 INFO::Fitting model to feature number 44, Coprobacter.fastidiosus
## 2024-07-16 13:50:59.992681 INFO::Fitting model to feature number 45, Coprococcus.catus
## 2024-07-16 13:50:59.994947 INFO::Fitting model to feature number 46, Coprococcus.comes
## 2024-07-16 13:50:59.997231 INFO::Fitting model to feature number 47, Coprococcus.eutactus
## 2024-07-16 13:50:59.999585 INFO::Fitting model to feature number 48, Dialister.invisus
## 2024-07-16 13:51:00.001825 INFO::Fitting model to feature number 49, Dielma.fastidiosa
## 2024-07-16 13:51:00.004077 INFO::Fitting model to feature number 50, Dorea.formicigenerans
## 2024-07-16 13:51:00.006367 INFO::Fitting model to feature number 51, Dorea.longicatena
## 2024-07-16 13:51:00.008667 INFO::Fitting model to feature number 52, Eggerthella.lenta
## 2024-07-16 13:51:00.010913 INFO::Fitting model to feature number 53, Eisenbergiella.massiliensis
## 2024-07-16 13:51:00.013177 INFO::Fitting model to feature number 54, Eisenbergiella.tayi
## 2024-07-16 13:51:00.015419 INFO::Fitting model to feature number 55, Enterocloster.aldenensis
## 2024-07-16 13:51:00.017728 INFO::Fitting model to feature number 56, Enterocloster.bolteae
## 2024-07-16 13:51:00.01999 INFO::Fitting model to feature number 57, Enterocloster.citroniae
## 2024-07-16 13:51:00.022213 INFO::Fitting model to feature number 58, Enterocloster.clostridioformis
## 2024-07-16 13:51:00.024488 INFO::Fitting model to feature number 59, Enterocloster.lavalensis
## 2024-07-16 13:51:00.026734 INFO::Fitting model to feature number 60, Erysipelatoclostridium.ramosum
## 2024-07-16 13:51:00.029009 INFO::Fitting model to feature number 61, Escherichia.coli
## 2024-07-16 13:51:00.031281 INFO::Fitting model to feature number 62, Eubacterium.ramulus
## 2024-07-16 13:51:00.033581 INFO::Fitting model to feature number 63, Eubacterium.sp..CAG.251
## 2024-07-16 13:51:00.035836 INFO::Fitting model to feature number 64, Eubacterium.sp..CAG.38
## 2024-07-16 13:51:00.038079 INFO::Fitting model to feature number 65, Eubacterium.ventriosum
## 2024-07-16 13:51:00.040396 INFO::Fitting model to feature number 66, Faecalibacterium.prausnitzii
## 2024-07-16 13:51:00.042633 INFO::Fitting model to feature number 67, Firmicutes.bacterium.CAG.83
## 2024-07-16 13:51:00.04494 INFO::Fitting model to feature number 68, Firmicutes.bacterium.CAG.94
## 2024-07-16 13:51:00.047171 INFO::Fitting model to feature number 69, Flavonifractor.plautii
## 2024-07-16 13:51:00.049552 INFO::Fitting model to feature number 70, Fusicatenibacter.saccharivorans
## 2024-07-16 13:51:00.05192 INFO::Fitting model to feature number 71, Gemmiger.formicilis
## 2024-07-16 13:51:00.054218 INFO::Fitting model to feature number 72, Haemophilus.parainfluenzae
## 2024-07-16 13:51:00.056506 INFO::Fitting model to feature number 73, Haemophilus.sp..HMSC71H05
## 2024-07-16 13:51:00.058769 INFO::Fitting model to feature number 74, Holdemania.filiformis
## 2024-07-16 13:51:00.061092 INFO::Fitting model to feature number 75, Hungatella.hathewayi
## 2024-07-16 13:51:00.063458 INFO::Fitting model to feature number 76, Intestinibacter.bartlettii
## 2024-07-16 13:51:00.065787 INFO::Fitting model to feature number 77, Intestinimonas.butyriciproducens
## 2024-07-16 13:51:00.068175 INFO::Fitting model to feature number 78, Klebsiella.pneumoniae
## 2024-07-16 13:51:00.070462 INFO::Fitting model to feature number 79, Klebsiella.variicola
## 2024-07-16 13:51:00.072822 INFO::Fitting model to feature number 80, Lachnospira.eligens
## 2024-07-16 13:51:00.075102 INFO::Fitting model to feature number 81, Lachnospira.pectinoschiza
## 2024-07-16 13:51:00.077425 INFO::Fitting model to feature number 82, Lacrimispora.saccharolytica
## 2024-07-16 13:51:00.079764 INFO::Fitting model to feature number 83, Lactobacillus.rogosae
## 2024-07-16 13:51:00.082057 INFO::Fitting model to feature number 84, Lawsonibacter.asaccharolyticus
## 2024-07-16 13:51:00.084365 INFO::Fitting model to feature number 85, Monoglobus.pectinilyticus
## 2024-07-16 13:51:00.086614 INFO::Fitting model to feature number 86, Odoribacter.splanchnicus
## 2024-07-16 13:51:00.08894 INFO::Fitting model to feature number 87, Oscillibacter.sp..57_20
## 2024-07-16 13:51:00.091216 INFO::Fitting model to feature number 88, Oscillibacter.sp..CAG.241
## 2024-07-16 13:51:00.093543 INFO::Fitting model to feature number 89, Parabacteroides.distasonis
## 2024-07-16 13:51:00.095838 INFO::Fitting model to feature number 90, Parabacteroides.merdae
## 2024-07-16 13:51:00.098127 INFO::Fitting model to feature number 91, Paraprevotella.clara
## 2024-07-16 13:51:00.100421 INFO::Fitting model to feature number 92, Paraprevotella.xylaniphila
## 2024-07-16 13:51:00.102694 INFO::Fitting model to feature number 93, Parasutterella.excrementihominis
## 2024-07-16 13:51:00.105001 INFO::Fitting model to feature number 94, Phascolarctobacterium.faecium
## 2024-07-16 13:51:00.107262 INFO::Fitting model to feature number 95, Phocaeicola.coprocola
## 2024-07-16 13:51:00.10957 INFO::Fitting model to feature number 96, Phocaeicola.dorei
## 2024-07-16 13:51:00.111847 INFO::Fitting model to feature number 97, Phocaeicola.massiliensis
## 2024-07-16 13:51:00.114107 INFO::Fitting model to feature number 98, Phocaeicola.plebeius
## 2024-07-16 13:51:00.11637 INFO::Fitting model to feature number 99, Phocaeicola.vulgatus
## 2024-07-16 13:51:00.118668 INFO::Fitting model to feature number 100, Prevotella.copri
## 2024-07-16 13:51:00.12096 INFO::Fitting model to feature number 101, Proteobacteria.bacterium.CAG.139
## 2024-07-16 13:51:00.123209 INFO::Fitting model to feature number 102, Roseburia.faecis
## 2024-07-16 13:51:00.125513 INFO::Fitting model to feature number 103, Roseburia.hominis
## 2024-07-16 13:51:00.127863 INFO::Fitting model to feature number 104, Roseburia.intestinalis
## 2024-07-16 13:51:00.130139 INFO::Fitting model to feature number 105, Roseburia.inulinivorans
## 2024-07-16 13:51:00.132431 INFO::Fitting model to feature number 106, Roseburia.sp..CAG.471
## 2024-07-16 13:51:00.134698 INFO::Fitting model to feature number 107, Ruminococcaceae.bacterium.D16
## 2024-07-16 13:51:00.137043 INFO::Fitting model to feature number 108, Ruminococcus.bicirculans
## 2024-07-16 13:51:00.139334 INFO::Fitting model to feature number 109, Ruminococcus.bromii
## 2024-07-16 13:51:00.141639 INFO::Fitting model to feature number 110, Ruthenibacterium.lactatiformans
## 2024-07-16 13:51:00.143922 INFO::Fitting model to feature number 111, Sellimonas.intestinalis
## 2024-07-16 13:51:00.146284 INFO::Fitting model to feature number 112, Streptococcus.parasanguinis
## 2024-07-16 13:51:00.148602 INFO::Fitting model to feature number 113, Streptococcus.salivarius
## 2024-07-16 13:51:00.150924 INFO::Fitting model to feature number 114, Turicimonas.muris
## 2024-07-16 13:51:00.153228 INFO::Fitting model to feature number 115, Veillonella.atypica
## 2024-07-16 13:51:00.155478 INFO::Fitting model to feature number 116, Veillonella.dispar
## 2024-07-16 13:51:00.15776 INFO::Fitting model to feature number 117, Veillonella.infantium
## 2024-07-16 13:51:00.160059 INFO::Fitting model to feature number 118, Veillonella.parvula
## 2024-07-16 13:51:00.162338 INFO::Fitting model to feature number 119, Veillonella.sp..T11011.6
## 2024-07-16 13:51:00.164609 INFO::Fitting model to feature number 120, X.Clostridium..innocuum
## 2024-07-16 13:51:00.166889 INFO::Fitting model to feature number 121, X.Clostridium..leptum
## 2024-07-16 13:51:00.169289 INFO::Fitting model to feature number 122, X.Clostridium..symbiosum
## 2024-07-16 13:51:00.171614 INFO::Fitting model to feature number 123, X.Eubacterium..rectale
## 2024-07-16 13:51:00.173973 INFO::Fitting model to feature number 124, X.Eubacterium..siraeum
## 2024-07-16 13:51:00.176291 INFO::Fitting model to feature number 125, X.Ruminococcus..gnavus
## 2024-07-16 13:51:00.178557 INFO::Fitting model to feature number 126, X.Ruminococcus..lactaris
## 2024-07-16 13:51:00.180865 INFO::Fitting model to feature number 127, X.Ruminococcus..torques
## 2024-07-16 13:51:00.201402 INFO::Counting total values for each feature
## 2024-07-16 13:51:00.212989 INFO::Writing filtered data to file output_species/features/filtered_data.tsv
## 2024-07-16 13:51:00.225485 INFO::Writing filtered, normalized data to file output_species/features/filtered_data_norm.tsv
## 2024-07-16 13:51:00.237504 INFO::Writing filtered, normalized, transformed data to file output_species/features/filtered_data_norm_transformed.tsv
## 2024-07-16 13:51:00.254926 INFO::Writing residuals to file output_species/fits/residuals.rds
## 2024-07-16 13:51:00.260917 INFO::Writing fitted values to file output_species/fits/fitted.rds
## 2024-07-16 13:51:00.266165 INFO::Writing all results to file (ordered by increasing q-values): output_species/all_results.tsv
## 2024-07-16 13:51:00.268997 INFO::Writing the significant results (those which are less than or equal to the threshold of 0.250000 ) to file (ordered by increasing q-values): output_species/significant_results.tsv
## 2024-07-16 13:51:00.269845 INFO::Writing heatmap of significant results to file: output_species/heatmap.pdf
## 2024-07-16 13:51:00.381671 INFO::Writing association plots (one for each significant association) to output folder: output_species
## 2024-07-16 13:51:00.384331 INFO::Plotting associations from most to least significant, grouped by metadata
## 2024-07-16 13:51:00.384864 INFO::Plotting data for metadata number 1, age
## 2024-07-16 13:51:00.385754 INFO::Creating scatter plot for continuous data, age vs Alistipes.indistinctus
## 2024-07-16 13:51:00.590621 INFO::Creating scatter plot for continuous data, age vs Lacrimispora.saccharolytica
## 2024-07-16 13:51:00.825569 INFO::Creating scatter plot for continuous data, age vs Ruminococcus.bicirculans
## 2024-07-16 13:51:00.94509 INFO::Creating scatter plot for continuous data, age vs Ruminococcus.bromii
## 2024-07-16 13:51:01.077986 INFO::Creating scatter plot for continuous data, age vs Bifidobacterium.pseudocatenulatum
## 2024-07-16 13:51:01.204905 INFO::Creating scatter plot for continuous data, age vs Haemophilus.parainfluenzae
## 2024-07-16 13:51:01.333034 INFO::Creating scatter plot for continuous data, age vs Lachnospira.eligens
## 2024-07-16 13:51:01.455966 INFO::Creating scatter plot for continuous data, age vs X.Ruminococcus..gnavus
## 2024-07-16 13:51:01.620915 INFO::Creating scatter plot for continuous data, age vs Butyricimonas.virosa
## 2024-07-16 13:51:01.741956 INFO::Creating scatter plot for continuous data, age vs Lawsonibacter.asaccharolyticus
## 2024-07-16 13:51:01.862373 INFO::Creating scatter plot for continuous data, age vs X.Eubacterium..siraeum
## 2024-07-16 13:51:01.985178 INFO::Creating scatter plot for continuous data, age vs Butyricimonas.synergistica
## 2024-07-16 13:51:02.107991 INFO::Creating scatter plot for continuous data, age vs Monoglobus.pectinilyticus
## 2024-07-16 13:51:02.229923 INFO::Creating scatter plot for continuous data, age vs Veillonella.atypica
## 2024-07-16 13:51:03.800608 INFO::Plotting data for metadata number 2, disease
## 2024-07-16 13:51:03.801811 INFO::Creating boxplot for categorical data, disease vs Alistipes.putredinis
## 2024-07-16 13:51:03.954377 INFO::Creating boxplot for categorical data, disease vs Gemmiger.formicilis
## 2024-07-16 13:51:04.081341 INFO::Creating boxplot for categorical data, disease vs X.Ruminococcus..torques
## 2024-07-16 13:51:04.195017 INFO::Creating boxplot for categorical data, disease vs Ruminococcus.bicirculans
## 2024-07-16 13:51:04.308347 INFO::Creating boxplot for categorical data, disease vs Sellimonas.intestinalis
## 2024-07-16 13:51:04.438 INFO::Creating boxplot for categorical data, disease vs X.Clostridium..leptum
## 2024-07-16 13:51:04.549976 INFO::Creating boxplot for categorical data, disease vs Alistipes.shahii
## 2024-07-16 13:51:05.624923 INFO::Plotting data for metadata number 3, antibiotics_current_use
## 2024-07-16 13:51:05.626108 INFO::Creating boxplot for categorical data, antibiotics_current_use vs Coprobacter.fastidiosus
## 2024-07-16 13:51:05.735708 INFO::Creating boxplot for categorical data, antibiotics_current_use vs X.Eubacterium..rectale
## 2024-07-16 13:51:05.846456 INFO::Creating boxplot for categorical data, antibiotics_current_use vs Agathobaculum.butyriciproducensUnlike gene expression studies, we do not have well-defined signatures or modules for microbiome data. Here, we will construct data-driven modules using weighted gene co-expression network analysis (WGCNA) (Langfelder and Horvath 2008), (Geistlinger et al. 2023). We aim to ensure that the effect of disease and other covariates has been removed by working on the residuals. Following the WGCNA tutorial, our first step will be to check whether there are any outliers in our data.
library(WGCNA)
datExpr <- as.data.frame(t(fit_data$residuals))
gsg = goodSamplesGenes(datExpr, verbose = 3)
## Flagging genes and samples with too many missing values...
## ..step 1
## ..Excluding 6 samples from the calculation due to too many missing genes.
## ..step 2
gsg$allOK
## [1] FALSEIf the last statement returns TRUE, no outliers are identified. If not, we need to remove the outliers from the data.
if (!gsg$allOK)
{if (sum(!gsg$goodGenes) > 0)
printFlush(paste("Removing genes:", paste(names(datExpr)[!gsg$goodGenes], collapse = ", ")));
if (sum(!gsg$goodSamples) > 0)
printFlush(paste("Removing samples:", paste(rownames(datExpr)[!gsg$goodSamples], collapse =", ")))
datExpr = datExpr[gsg$goodSamples, gsg$goodGenes]
}
## Removing samples: CSM5MCVB_P, CSM79HNY_P, ESM5GEYY_P, ESM718U9_P, MSM6J2N6_P, MSM9VZLX_PAfter removing the outliers, we need to choose a suitable soft threshold parameter for creating the modules as part of the WGCNA algorithm. This power value must produce a graph similar to a scale-free network. We can use the mean connectivity graphic for the selection of this power parameter.
# Choose a set of soft threshold parameters
powers = c(c(1:20), seq(from = 22, to=30, by=2))
sft = pickSoftThreshold(datExpr, powerVector = powers, verbose = 5, dataIsExpr = TRUE, RsquaredCut = 0.30)
## pickSoftThreshold: will use block size 127.
## pickSoftThreshold: calculating connectivity for given powers...
## ..working on genes 1 through 127 of 127
## Power SFT.R.sq slope truncated.R.sq mean.k. median.k. max.k.
## 1 1 0.1790 -0.972 0.85700 15.00000 1.48e+01 23.0000
## 2 2 0.3450 -0.729 0.63500 2.95000 2.85e+00 5.6900
## 3 3 0.2400 -4.130 0.12700 0.80400 7.36e-01 2.6100
## 4 4 0.3100 -4.870 0.19900 0.29200 2.14e-01 1.6700
## 5 5 0.2150 -4.030 -0.00880 0.13700 7.02e-02 1.2200
## 6 6 0.1800 -2.930 -0.05130 0.07910 2.46e-02 0.9570
## 7 7 0.1380 -2.240 -0.10100 0.05320 9.33e-03 0.7900
## 8 8 0.2200 -2.790 -0.00259 0.03940 3.71e-03 0.6690
## 9 9 0.0872 -1.450 -0.09130 0.03100 1.50e-03 0.5760
## 10 10 0.1190 -1.630 -0.07660 0.02540 6.73e-04 0.5020
## 11 11 0.0562 -1.250 -0.03470 0.02130 2.66e-04 0.4410
## 12 12 0.1620 -2.150 -0.07720 0.01810 1.12e-04 0.3910
## 13 13 0.2120 -2.350 -0.00949 0.01560 4.84e-05 0.3480
## 14 14 0.2480 -2.410 0.04950 0.01360 2.12e-05 0.3110
## 15 15 0.2890 -2.470 0.15200 0.01190 9.53e-06 0.2810
## 16 16 0.3030 -2.420 0.18400 0.01050 4.33e-06 0.2580
## 17 17 0.2410 -2.380 0.02450 0.00933 1.98e-06 0.2380
## 18 18 0.3190 -2.640 0.15400 0.00830 9.03e-07 0.2180
## 19 19 0.3320 -2.600 0.17700 0.00741 4.03e-07 0.2010
## 20 20 0.2370 -2.050 0.02180 0.00664 1.70e-07 0.1840
## 21 22 0.2350 -2.000 0.07520 0.00536 3.40e-08 0.1560
## 22 24 0.2430 -2.230 0.09560 0.00437 6.98e-09 0.1310
## 23 26 0.2370 -2.120 0.26200 0.00358 1.44e-09 0.1110
## 24 28 0.2490 -2.080 0.26200 0.00294 2.93e-10 0.0937
## 25 30 0.2490 -2.290 0.30100 0.00243 5.67e-11 0.0791In this step, we will conduct a one-step module detection based on the selected soft threshold parameter selected above.
power = sft$powerEstimate
net = blockwiseModules(datExpr,
power = power,
corFnc="bicor",
corOptions=list(maxPOutliers=0.1),
networkType ="unsigned",
maxBlockSize = ncol(datExpr),
minModuleSize = 3,
TOMType = "unsigned",
reassignThreshold = 0,
mergeCutHeight = 0,
verbose = 3)
## Calculating module eigengenes block-wise from all genes
## Flagging genes and samples with too many missing values...
## ..step 1
## ..Working on block 1 .
## TOM calculation: adjacency..
## ..will not use multithreading.
## Fraction of slow calculations: 0.000000
## ..connectivity..
## ..matrix multiplication (system BLAS)..
## ..normalization..
## ..done.
## ....clustering..
## ....detecting modules..
## ....calculating module eigengenes..
## ....checking kME in modules..
## ..removing 3 genes from module 1 because their KME is too low.
## ..removing 3 genes from module 2 because their KME is too low.
## ..removing 2 genes from module 3 because their KME is too low.
## ..removing 1 genes from module 5 because their KME is too low.
## ..removing 1 genes from module 7 because their KME is too low.
## ..merging modules that are too close..
## mergeCloseModules: Merging modules whose distance is less than 0
## Calculating new MEs...
####################
# How many modules #
####################
ncol(net$MEs)
## [1] 14
table(net$colors)
##
## black blue brown green greenyellow grey
## 7 14 13 12 5 10
## magenta pink purple red salmon tan
## 6 6 5 11 3 5
## turquoise yellow
## 18 12The WGCNA algorithm produced 14 modules which we can visualize as follows.
##########################
# Plot module dendrogram #
##########################
eigenGenes <- net$MEs
MEDiss = 1-cor(eigenGenes)
METree = hclust(as.dist(MEDiss), method = "average")
plot(METree, main = "Clustering of module eigengenes", xlab = "", sub = "")
Next, we calculate hub genes for the modules and create the mapping files to proceed with the MSEA.
###########################################
# Re-calculate modules and find hub genes #
###########################################
moduleColors <- net$colors
MEs0 = moduleEigengenes(datExpr, moduleColors)$eigengenes
modules_data = orderMEs(MEs0)
#######################
# Create mapping file #
#######################
library(tidyverse)
feature_by_modules <- as.data.frame(net$colors)
feature_by_modules <- rownames_to_column(feature_by_modules)
colnames(feature_by_modules) <- c('Feature', 'Module')
features_mapping <- feature_by_modules
features_mapping$Module <- paste('ME', features_mapping$Module, sep = '')Finally, we will run the MSEA analysis on the modules we constructed using WGCNA. Here, we first create a wrapper for the MSEA analysis using the gsEasy package.
library(reshape2)
library(gsEasy)
################
# MSEA Wrapper #
################
run_MSEA <- function(microbeSet, # A list
ranked_features, # Ranked list of featured
filter.count = 3,
seed = 1234,
fdr.correction = 'BH') {
###################
# Filter out sets #
##################
microbeSet0 <- microbeSet
cond <- sapply(microbeSet0, function(x) length(x) > filter.count)
microbeSet <- microbeSet0[cond]
lengthmicrobeSet <- as.data.frame(reshape2::melt(lapply(microbeSet, function(x) length(x))))
colnames(lengthmicrobeSet) <- c('Freq','Set')
################
# Classic MSEA #
################
set.seed(seed)
enrichment <- as.data.frame(sapply(microbeSet, function(set) gset(S = set, r = ranked_features)))
colnames(enrichment)<-'ES'
enrichment <- rownames_to_column(enrichment, 'Set')
enrichment <- merge(enrichment, lengthmicrobeSet, 'Set')
enrichment$qval <- p.adjust(enrichment$ES, fdr.correction)
##########
# Return #
##########
return(enrichment)
}Before running the MSEA, we also need to rank the differential analysis results from MaAsLin2. We use the topGo package to create a list of microbe sets from the mapping file created above.
###################
# Rank DA results #
###################
results <- fit_data$results %>% filter(metadata == 'disease')
results$qval <- p.adjust(results$pval, 'BH')
results <- results[order(results$qval, decreasing = FALSE), ]
###################
# MSEA Processing #
###################
library(topGO)
module_map <- features_mapping
mod.gs <- tapply(module_map$Module, module_map$Feature, as.character)
microbeSet <- inverseList(mod.gs)
microbeSet
## $MEblack
## [1] "Anaeromassilibacillus.sp..An250" "Anaerotruncus.colihominis"
## [3] "Blautia.wexlerae" "Eisenbergiella.tayi"
## [5] "Firmicutes.bacterium.CAG.94" "Ruthenibacterium.lactatiformans"
## [7] "Sellimonas.intestinalis"
##
## $MEblue
## [1] "Clostridium.sp..CAG.58" "Erysipelatoclostridium.ramosum"
## [3] "Haemophilus.parainfluenzae" "Intestinibacter.bartlettii"
## [5] "Klebsiella.pneumoniae" "Klebsiella.variicola"
## [7] "Lawsonibacter.asaccharolyticus" "Streptococcus.parasanguinis"
## [9] "Streptococcus.salivarius" "Veillonella.atypica"
## [11] "Veillonella.dispar" "Veillonella.infantium"
## [13] "Veillonella.parvula" "Veillonella.sp..T11011.6"
##
## $MEbrown
## [1] "Alistipes.finegoldii" "Alistipes.indistinctus"
## [3] "Alistipes.putredinis" "Alistipes.shahii"
## [5] "Bacteroides.xylanisolvens" "Bilophila.wadsworthia"
## [7] "Firmicutes.bacterium.CAG.83" "Odoribacter.splanchnicus"
## [9] "Oscillibacter.sp..57_20" "Oscillibacter.sp..CAG.241"
## [11] "Phocaeicola.dorei" "Ruminococcus.bromii"
## [13] "X.Eubacterium..siraeum"
##
## $MEgreen
## [1] "Blautia.sp..CAG.257" "Clostridium.bolteae.CAG.59"
## [3] "Eggerthella.lenta" "Eisenbergiella.massiliensis"
## [5] "Enterocloster.aldenensis" "Enterocloster.bolteae"
## [7] "Enterocloster.citroniae" "Enterocloster.clostridioformis"
## [9] "Enterocloster.lavalensis" "Flavonifractor.plautii"
## [11] "Lacrimispora.saccharolytica" "X.Clostridium..symbiosum"
##
## $MEgreenyellow
## [1] "Coprobacter.fastidiosus" "Escherichia.coli"
## [3] "Hungatella.hathewayi" "X.Clostridium..innocuum"
## [5] "X.Ruminococcus..gnavus"
##
## $MEgrey
## [1] "Anaerotignum.lactatifermentans" "Bacteroides.cellulosilyticus"
## [3] "Bacteroides.fragilis" "Collinsella.intestinalis"
## [5] "Eubacterium.sp..CAG.38" "Haemophilus.sp..HMSC71H05"
## [7] "Intestinimonas.butyriciproducens" "Lachnospira.eligens"
## [9] "Roseburia.intestinalis" "Roseburia.sp..CAG.471"
##
## $MEmagenta
## [1] "Bacteroides.finegoldii" "Paraprevotella.clara"
## [3] "Paraprevotella.xylaniphila" "Phocaeicola.coprocola"
## [5] "Phocaeicola.plebeius" "Prevotella.copri"
##
## $MEpink
## [1] "Bacteroides.galacturonicus" "Bifidobacterium.pseudocatenulatum"
## [3] "Eubacterium.sp..CAG.251" "Lachnospira.pectinoschiza"
## [5] "Lactobacillus.rogosae" "Phocaeicola.massiliensis"
##
## $MEpurple
## [1] "Barnesiella.intestinihominis" "Butyricimonas.synergistica"
## [3] "Butyricimonas.virosa" "Coprococcus.eutactus"
## [5] "Ruminococcus.bicirculans"
##
## $MEred
## [1] "Akkermansia.muciniphila" "Bacteroides.intestinalis"
## [3] "Clostridium.sp..CAG.299" "Dialister.invisus"
## [5] "Dielma.fastidiosa" "Holdemania.filiformis"
## [7] "Monoglobus.pectinilyticus" "Parasutterella.excrementihominis"
## [9] "Proteobacteria.bacterium.CAG.139" "Turicimonas.muris"
## [11] "X.Clostridium..leptum"
##
## $MEsalmon
## [1] "Bacteroides.faecis" "Bacteroides.faecis.CAG.32"
## [3] "Phascolarctobacterium.faecium"
##
## $MEtan
## [1] "Bacteroides.salyersiae" "Bifidobacterium.adolescentis"
## [3] "Bifidobacterium.bifidum" "Bifidobacterium.longum"
## [5] "Collinsella.aerofaciens"
##
## $MEturquoise
## [1] "Agathobaculum.butyriciproducens" "Anaerobutyricum.hallii"
## [3] "Anaerostipes.hadrus" "Bacteroides.eggerthii"
## [5] "Blautia.obeum" "Coprococcus.catus"
## [7] "Coprococcus.comes" "Dorea.formicigenerans"
## [9] "Dorea.longicatena" "Eubacterium.ramulus"
## [11] "Faecalibacterium.prausnitzii" "Fusicatenibacter.saccharivorans"
## [13] "Gemmiger.formicilis" "Roseburia.faecis"
## [15] "Roseburia.hominis" "Roseburia.inulinivorans"
## [17] "X.Eubacterium..rectale" "X.Ruminococcus..torques"
##
## $MEyellow
## [1] "Acidaminococcus.intestini" "Bacteroides.caccae"
## [3] "Bacteroides.ovatus" "Bacteroides.stercoris"
## [5] "Bacteroides.thetaiotaomicron" "Bacteroides.uniformis"
## [7] "Eubacterium.ventriosum" "Parabacteroides.distasonis"
## [9] "Parabacteroides.merdae" "Phocaeicola.vulgatus"
## [11] "Ruminococcaceae.bacterium.D16" "X.Ruminococcus..lactaris"We are now ready to run the MSEA analysis. We run \(100,000\) permutations to calculate the enrichment scores.
We can plot the enrichment scores to visualize the MSEA results.
p <- MSEA %>%
arrange(-pval) %>%
mutate(ID = factor(ID, levels = ID)) %>%
ggplot(aes(y = -log10(pval), x = ID)) +
geom_bar(stat = "identity", fill = 'cornflowerblue') + theme_bw() +
coord_flip() +
ggtitle('Statistically significant modules associated with disease') +
xlab('') +
ylab('MSEA enrichment score')
print(p)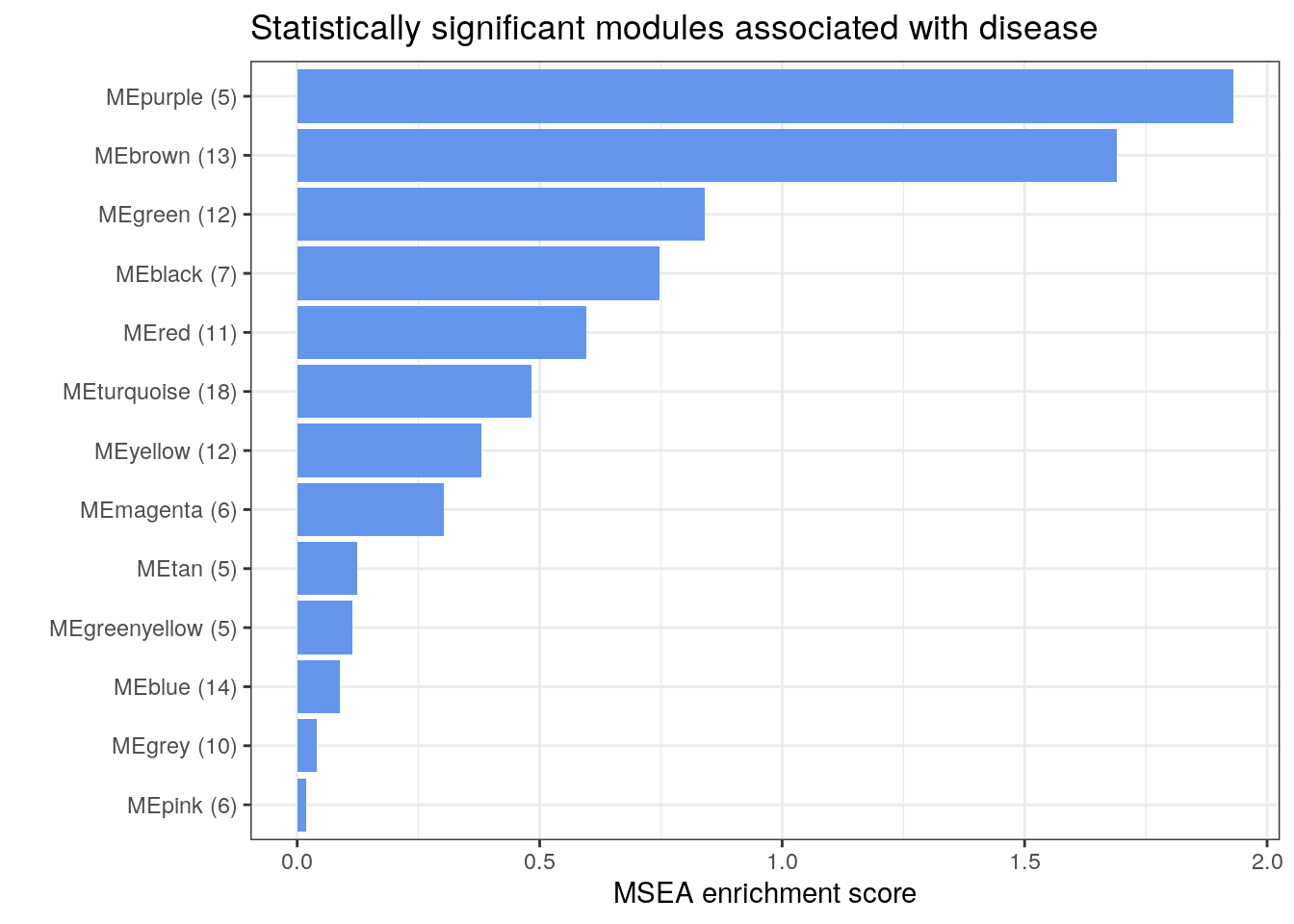
Based on the MSEA results, we obtain 13 enriched modules of microbial species. We can also examine the members of the top enriched modules.
17.3 Print the most significant modules for species relative abundance data
microbeSet[["MEpurple"]]
## [1] "Barnesiella.intestinihominis" "Butyricimonas.synergistica"
## [3] "Butyricimonas.virosa" "Coprococcus.eutactus"
## [5] "Ruminococcus.bicirculans"
microbeSet[["MEbrown"]]
## [1] "Alistipes.finegoldii" "Alistipes.indistinctus"
## [3] "Alistipes.putredinis" "Alistipes.shahii"
## [5] "Bacteroides.xylanisolvens" "Bilophila.wadsworthia"
## [7] "Firmicutes.bacterium.CAG.83" "Odoribacter.splanchnicus"
## [9] "Oscillibacter.sp..57_20" "Oscillibacter.sp..CAG.241"
## [11] "Phocaeicola.dorei" "Ruminococcus.bromii"
## [13] "X.Eubacterium..siraeum"17.4 Performing the MSEA analysis with pathway relative abundance data
Next, we repeat the MSEA with the pathway relative abundance data from the iHMP project and follow the same steps as before.
##########################
# Load HMP2 pathway data #
##########################
se_pathway <- sampleMetadata |>
filter(study_name == "HMP_2019_ibdmdb") |>
returnSamples("pathway_abundance", rownames = "short")
##########################
# Create sample metadata #
##########################
sample_metadata <-
colData(se_pathway) %>%
as.data.frame() %>% filter(visit_number == 1) %>%
dplyr::select("age", "disease", "antibiotics_current_use")
# Set reference
sample_metadata$disease <- as.factor(sample_metadata$disease)
sample_metadata$disease <- relevel(sample_metadata$disease, 'healthy')
###########################
# Create Pathway Features #
###########################
feature_pwys_t <- as.data.frame(assay(se_pathway))
feature_pwys_t <- rownames_to_column(feature_pwys_t, "ID")
feature_pwys_t <- feature_pwys_t %>%
filter(!grepl("\\|", ID)) %>%
filter(!ID %in% c('UNMAPPED', 'UNINTEGRATED')) %>%
column_to_rownames('ID') %>%
as.data.frame()
##############################
# Subset to baseline samples #
##############################
feature_pwys <- as.data.frame(t(feature_pwys_t))
feature_pwys <- feature_pwys[rownames(sample_metadata),]
feature_pwys <- feature_pwys / 100
rm(feature_pwys_t); rm(se_pathway)As before, we first run a MaAsLin2 analysis using default settings and construct the modules using residuals from the MaAsLin2 models.
fit_data = Maaslin2(input_data = feature_pwys,
input_metadata = sample_metadata,
normalization = "NONE",
output = "output_pwys",
fixed_effects = c("disease", "age", "antibiotics_current_use"))
## [1] "Creating output folder"
## [1] "Creating output feature tables folder"
## [1] "Creating output fits folder"
## [1] "Creating output figures folder"
## 2024-07-16 13:51:29.224589 INFO::Writing function arguments to log file
## 2024-07-16 13:51:29.228387 INFO::Verifying options selected are valid
## 2024-07-16 13:51:29.228854 INFO::Determining format of input files
## 2024-07-16 13:51:29.229307 INFO::Input format is data samples as rows and metadata samples as rows
## 2024-07-16 13:51:29.234694 INFO::Formula for fixed effects: expr ~ disease + age + antibiotics_current_use
## 2024-07-16 13:51:29.235236 INFO::Factor detected for categorial metadata 'disease'. Provide a reference argument or manually set factor ordering to change reference level.
## 2024-07-16 13:51:29.235859 INFO::Filter data based on min abundance and min prevalence
## 2024-07-16 13:51:29.236292 INFO::Total samples in data: 136
## 2024-07-16 13:51:29.236706 INFO::Min samples required with min abundance for a feature not to be filtered: 13.600000
## 2024-07-16 13:51:29.242046 INFO::Total filtered features: 113
## 2024-07-16 13:51:29.242615 INFO::Filtered feature names from abundance and prevalence filtering: PWY.5044..purine.nucleotides.degradation.I..plants., PROPFERM.PWY..L.alanine.fermentation.to.propanoate.and.acetate, PWY.6596..adenosine.nucleotides.degradation.I, PWY.5004..superpathway.of.L.citrulline.metabolism, UDPNACETYLGALSYN.PWY..UDP.N.acetyl.D.glucosamine.biosynthesis.II, PWY66.367..ketogenesis, PWY.5392..reductive.TCA.cycle.II, PWY.101..photosynthesis.light.reactions, PWY.7031..protein.N.glycosylation..bacterial., PWY.5265..peptidoglycan.biosynthesis.II..staphylococci., PWY.7268..NAD.NADP.NADH.NADPH.cytosolic.interconversion..yeast., PWY.7165..L.ascorbate.biosynthesis.VI..engineered.pathway., CATECHOL.ORTHO.CLEAVAGE.PWY..catechol.degradation.to..beta..ketoadipate, PWY.5417..catechol.degradation.III..ortho.cleavage.pathway., PWY.5431..aromatic.compounds.degradation.via..beta..ketoadipate, PWY3DJ.35471..L.ascorbate.biosynthesis.IV, PWY.6185..4.methylcatechol.degradation..ortho.cleavage., PWY.7245..superpathway.NAD.NADP...NADH.NADPH.interconversion..yeast., PWY.5181..toluene.degradation.III..aerobic...via.p.cresol., PWY.6562..norspermidine.biosynthesis, PWY.7431..aromatic.biogenic.amine.degradation..bacteria., PWY.6307..L.tryptophan.degradation.X..mammalian..via.tryptamine., PWY.6313..serotonin.degradation, URSIN.PWY..ureide.biosynthesis, LIPASYN.PWY..phospholipases, DHGLUCONATE.PYR.CAT.PWY..glucose.degradation..oxidative., DENITRIFICATION.PWY..nitrate.reduction.I..denitrification., PWY.6662..superpathway.of.quinolone.and.alkylquinolone.biosynthesis, PWY.6660..2.heptyl.3.hydroxy.4.1H..quinolone.biosynthesis, PWY.6182..superpathway.of.salicylate.degradation, P165.PWY..superpathway.of.purines.degradation.in.plants, PWY66.388..fatty.acid..alpha..oxidation.III, TYRFUMCAT.PWY..L.tyrosine.degradation.I, PWY.5101..L.isoleucine.biosynthesis.II, PWY.6167..flavin.biosynthesis.II..archaea., PWY.6748..nitrate.reduction.VII..denitrification., PWY.7039..phosphatidate.metabolism..as.a.signaling.molecule, VALDEG.PWY..L.valine.degradation.I, PWY.5198..factor.420.biosynthesis, PWY.6215..4.chlorobenzoate.degradation, AEROBACTINSYN.PWY..aerobactin.biosynthesis, P562.PWY..myo.inositol.degradation.I, PWY.7409..phospholipid.remodeling..phosphatidylethanolamine..yeast., PWY.4722..creatinine.degradation.II, PWY.3801..sucrose.degradation.II..sucrose.synthase., PWY.7345..superpathway.of.anaerobic.sucrose.degradation, P125.PWY..superpathway.of..R.R..butanediol.biosynthesis, PWY.5994..palmitate.biosynthesis.I..animals.and.fungi., PWY.4321..L.glutamate.degradation.IV, PWY.7528..L.methionine.salvage.cycle.I..bacteria.and.plants., PWY.4361..S.methyl.5.thio..alpha..D.ribose.1.phosphate.degradation, PWY.1541..superpathway.of.taurine.degradation, PWY.5654..2.amino.3.carboxymuconate.semialdehyde.degradation.to.2.oxopentenoate, PWY.6210..2.aminophenol.degradation, PWY.6071..superpathway.of.phenylethylamine.degradation, PWY0.321..phenylacetate.degradation.I..aerobic., PWY.7200..superpathway.of.pyrimidine.deoxyribonucleoside.salvage, PWY.2221..Entner.Doudoroff.pathway.III..semi.phosphorylative., PWY.6992..1.5.anhydrofructose.degradation, PWY.7294..xylose.degradation.IV, PWY.6145..superpathway.of.sialic.acids.and.CMP.sialic.acids.biosynthesis, PWY.5180..toluene.degradation.I..aerobic...via.o.cresol., PWY.5182..toluene.degradation.II..aerobic...via.4.methylcatechol., PWY.5415..catechol.degradation.I..meta.cleavage.pathway., PWY.6785..hydrogen.production.VIII, PWY.5509..adenosylcobalamin.biosynthesis.from.cobyrinate.a.c.diamide.I, PWY.6641..superpathway.of.sulfolactate.degradation, PWY.7527..L.methionine.salvage.cycle.III, PWY.6396..superpathway.of.2.3.butanediol.biosynthesis, PWY.6467..Kdo.transfer.to.lipid.IVA.III..Chlamydia., X7ALPHADEHYDROX.PWY..cholate.degradation..bacteria..anaerobic., PWY.7374..1.4.dihydroxy.6.naphthoate.biosynthesis.I, PWY.6906..chitin.derivatives.degradation, PWY.5514..UDP.N.acetyl.D.galactosamine.biosynthesis.II, PWY.7317..superpathway.of.dTDP.glucose.derived.O.antigen.building.blocks.biosynthesis, PWY.7218..photosynthetic.3.hydroxybutanoate.biosynthesis..engineered., PHOTOALL.PWY..oxygenic.photosynthesis, PWY.6344..L.ornithine.degradation.II..Stickland.reaction., P621.PWY..nylon.6.oligomer.degradation, PWY.5028..L.histidine.degradation.II, CRNFORCAT.PWY..creatinine.degradation.I, PWY.6269..adenosylcobalamin.salvage.from.cobinamide.II, PWY.7389..superpathway.of.anaerobic.energy.metabolism..invertebrates., PWY.7384..anaerobic.energy.metabolism..invertebrates..mitochondrial., PWY.6165..chorismate.biosynthesis.II..archaea., PWY5F9.12..biphenyl.degradation, PWY.5647..2.nitrobenzoate.degradation.I, PWY.6138..CMP.N.acetylneuraminate.biosynthesis.I..eukaryotes., PWY.5910..superpathway.of.geranylgeranyldiphosphate.biosynthesis.I..via.mevalonate., PWY.922..mevalonate.pathway.I, PWY.6435..4.hydroxybenzoate.biosynthesis.V, PWY3O.1109..superpathway.of.4.hydroxybenzoate.biosynthesis..yeast., PWY.5754..4.hydroxybenzoate.biosynthesis.I..eukaryotes., PWY1G.0..mycothiol.biosynthesis, PWY.1501..mandelate.degradation.I, PWY.6107..chlorosalicylate.degradation, PWY.5534..propylene.degradation, PWY.7118..chitin.degradation.to.ethanol, PWY.7290..Escherichia.coli.serotype.O86.O.antigen.biosynthesis, PWY.181..photorespiration, PWY.1622..formaldehyde.assimilation.I..serine.pathway., CODH.PWY..reductive.acetyl.coenzyme.A.pathway, PWY.6349..CDP.archaeol.biosynthesis, PWY.6174..mevalonate.pathway.II..archaea., PWY.7286..7..3.amino.3.carboxypropyl..wyosine.biosynthesis, METHANOGENESIS.PWY..methanogenesis.from.H2.and.CO2, PWY.7391..isoprene.biosynthesis.II..engineered., PWY.5055..nicotinate.degradation.III, PWY.7399..methylphosphonate.degradation.II, P241.PWY..coenzyme.B.biosynthesis, PWY.5178..toluene.degradation.IV..aerobic...via.catechol., PWY.5420..catechol.degradation.II..meta.cleavage.pathway., PWY.5419..catechol.degradation.to.2.oxopent.4.enoate.II
## 2024-07-16 13:51:29.248828 INFO::Total filtered features with variance filtering: 0
## 2024-07-16 13:51:29.24933 INFO::Filtered feature names from variance filtering:
## 2024-07-16 13:51:29.249761 INFO::Running selected normalization method: NONE
## 2024-07-16 13:51:29.250172 INFO::Applying z-score to standardize continuous metadata
## 2024-07-16 13:51:29.253589 INFO::Running selected transform method: LOG
## 2024-07-16 13:51:29.261319 INFO::Running selected analysis method: LM
## 2024-07-16 13:51:29.261865 INFO::Fitting model to feature number 1, PWY.1042..glycolysis.IV..plant.cytosol.
## 2024-07-16 13:51:29.264535 INFO::Fitting model to feature number 2, DTDPRHAMSYN.PWY..dTDP.L.rhamnose.biosynthesis.I
## 2024-07-16 13:51:29.266852 INFO::Fitting model to feature number 3, PWY.5695..urate.biosynthesis.inosine.5..phosphate.degradation
## 2024-07-16 13:51:29.269177 INFO::Fitting model to feature number 4, PWY.6936..seleno.amino.acid.biosynthesis
## 2024-07-16 13:51:29.271458 INFO::Fitting model to feature number 5, ILEUSYN.PWY..L.isoleucine.biosynthesis.I..from.threonine.
## 2024-07-16 13:51:29.273771 INFO::Fitting model to feature number 6, PWY.7111..pyruvate.fermentation.to.isobutanol..engineered.
## 2024-07-16 13:51:29.2761 INFO::Fitting model to feature number 7, VALSYN.PWY..L.valine.biosynthesis
## 2024-07-16 13:51:29.278378 INFO::Fitting model to feature number 8, PWY.6609..adenine.and.adenosine.salvage.III
## 2024-07-16 13:51:29.280673 INFO::Fitting model to feature number 9, PWY.6737..starch.degradation.V
## 2024-07-16 13:51:29.282971 INFO::Fitting model to feature number 10, PWY.7219..adenosine.ribonucleotides.de.novo.biosynthesis
## 2024-07-16 13:51:29.285283 INFO::Fitting model to feature number 11, PWY0.1586..peptidoglycan.maturation..meso.diaminopimelate.containing.
## 2024-07-16 13:51:29.28758 INFO::Fitting model to feature number 12, ASPASN.PWY..superpathway.of.L.aspartate.and.L.asparagine.biosynthesis
## 2024-07-16 13:51:29.289872 INFO::Fitting model to feature number 13, PWY.5973..cis.vaccenate.biosynthesis
## 2024-07-16 13:51:29.292217 INFO::Fitting model to feature number 14, PWY.7221..guanosine.ribonucleotides.de.novo.biosynthesis
## 2024-07-16 13:51:29.294508 INFO::Fitting model to feature number 15, PWY.5686..UMP.biosynthesis
## 2024-07-16 13:51:29.296832 INFO::Fitting model to feature number 16, X1CMET2.PWY..N10.formyl.tetrahydrofolate.biosynthesis
## 2024-07-16 13:51:29.299129 INFO::Fitting model to feature number 17, PWY.6122..5.aminoimidazole.ribonucleotide.biosynthesis.II
## 2024-07-16 13:51:29.301425 INFO::Fitting model to feature number 18, PWY.6277..superpathway.of.5.aminoimidazole.ribonucleotide.biosynthesis
## 2024-07-16 13:51:29.303747 INFO::Fitting model to feature number 19, PWY.6385..peptidoglycan.biosynthesis.III..mycobacteria.
## 2024-07-16 13:51:29.306017 INFO::Fitting model to feature number 20, PWY.3841..folate.transformations.II
## 2024-07-16 13:51:29.308345 INFO::Fitting model to feature number 21, RIBOSYN2.PWY..flavin.biosynthesis.I..bacteria.and.plants.
## 2024-07-16 13:51:29.310642 INFO::Fitting model to feature number 22, PWY.6387..UDP.N.acetylmuramoyl.pentapeptide.biosynthesis.I..meso.diaminopimelate.containing.
## 2024-07-16 13:51:29.312949 INFO::Fitting model to feature number 23, THISYNARA.PWY..superpathway.of.thiamin.diphosphate.biosynthesis.III..eukaryotes.
## 2024-07-16 13:51:29.315235 INFO::Fitting model to feature number 24, PWY.7663..gondoate.biosynthesis..anaerobic.
## 2024-07-16 13:51:29.317555 INFO::Fitting model to feature number 25, PWY.6386..UDP.N.acetylmuramoyl.pentapeptide.biosynthesis.II..lysine.containing.
## 2024-07-16 13:51:29.31989 INFO::Fitting model to feature number 26, PWY.6700..queuosine.biosynthesis
## 2024-07-16 13:51:29.322341 INFO::Fitting model to feature number 27, PEPTIDOGLYCANSYN.PWY..peptidoglycan.biosynthesis.I..meso.diaminopimelate.containing.
## 2024-07-16 13:51:29.324677 INFO::Fitting model to feature number 28, TRNA.CHARGING.PWY..tRNA.charging
## 2024-07-16 13:51:29.326949 INFO::Fitting model to feature number 29, PWY.6121..5.aminoimidazole.ribonucleotide.biosynthesis.I
## 2024-07-16 13:51:29.329274 INFO::Fitting model to feature number 30, HISTSYN.PWY..L.histidine.biosynthesis
## 2024-07-16 13:51:29.331601 INFO::Fitting model to feature number 31, PWY.7229..superpathway.of.adenosine.nucleotides.de.novo.biosynthesis.I
## 2024-07-16 13:51:29.333906 INFO::Fitting model to feature number 32, PWY.7199..pyrimidine.deoxyribonucleosides.salvage
## 2024-07-16 13:51:29.336192 INFO::Fitting model to feature number 33, PANTO.PWY..phosphopantothenate.biosynthesis.I
## 2024-07-16 13:51:29.338476 INFO::Fitting model to feature number 34, PWY.2942..L.lysine.biosynthesis.III
## 2024-07-16 13:51:29.340862 INFO::Fitting model to feature number 35, PWY.7237..myo...chiro..and.scillo.inositol.degradation
## 2024-07-16 13:51:29.343173 INFO::Fitting model to feature number 36, PWY.6168..flavin.biosynthesis.III..fungi.
## 2024-07-16 13:51:29.345489 INFO::Fitting model to feature number 37, COA.PWY.1..coenzyme.A.biosynthesis.II..mammalian.
## 2024-07-16 13:51:29.347835 INFO::Fitting model to feature number 38, PWY.5667..CDP.diacylglycerol.biosynthesis.I
## 2024-07-16 13:51:29.350123 INFO::Fitting model to feature number 39, PWY0.1319..CDP.diacylglycerol.biosynthesis.II
## 2024-07-16 13:51:29.352435 INFO::Fitting model to feature number 40, PWY.5097..L.lysine.biosynthesis.VI
## 2024-07-16 13:51:29.354694 INFO::Fitting model to feature number 41, ANAGLYCOLYSIS.PWY..glycolysis.III..from.glucose.
## 2024-07-16 13:51:29.357022 INFO::Fitting model to feature number 42, PWY.6123..inosine.5..phosphate.biosynthesis.I
## 2024-07-16 13:51:29.359312 INFO::Fitting model to feature number 43, ARGININE.SYN4.PWY..L.ornithine.de.novo..biosynthesis
## 2024-07-16 13:51:29.361605 INFO::Fitting model to feature number 44, PWY.6163..chorismate.biosynthesis.from.3.dehydroquinate
## 2024-07-16 13:51:29.363916 INFO::Fitting model to feature number 45, THRESYN.PWY..superpathway.of.L.threonine.biosynthesis
## 2024-07-16 13:51:29.366189 INFO::Fitting model to feature number 46, PYRIDNUCSYN.PWY..NAD.biosynthesis.I..from.aspartate.
## 2024-07-16 13:51:29.368491 INFO::Fitting model to feature number 47, PWY.6124..inosine.5..phosphate.biosynthesis.II
## 2024-07-16 13:51:29.370752 INFO::Fitting model to feature number 48, PWY.6147..6.hydroxymethyl.dihydropterin.diphosphate.biosynthesis.I
## 2024-07-16 13:51:29.373057 INFO::Fitting model to feature number 49, PWY.7539..6.hydroxymethyl.dihydropterin.diphosphate.biosynthesis.III..Chlamydia.
## 2024-07-16 13:51:29.375331 INFO::Fitting model to feature number 50, PWY.6151..S.adenosyl.L.methionine.cycle.I
## 2024-07-16 13:51:29.377616 INFO::Fitting model to feature number 51, SER.GLYSYN.PWY..superpathway.of.L.serine.and.glycine.biosynthesis.I
## 2024-07-16 13:51:29.379921 INFO::Fitting model to feature number 52, PWY.6126..superpathway.of.adenosine.nucleotides.de.novo.biosynthesis.II
## 2024-07-16 13:51:29.382178 INFO::Fitting model to feature number 53, PANTOSYN.PWY..pantothenate.and.coenzyme.A.biosynthesis.I
## 2024-07-16 13:51:29.384468 INFO::Fitting model to feature number 54, PWY.7228..superpathway.of.guanosine.nucleotides.de.novo.biosynthesis.I
## 2024-07-16 13:51:29.386732 INFO::Fitting model to feature number 55, COA.PWY..coenzyme.A.biosynthesis.I
## 2024-07-16 13:51:29.389036 INFO::Fitting model to feature number 56, PWY.4242..pantothenate.and.coenzyme.A.biosynthesis.III
## 2024-07-16 13:51:29.391304 INFO::Fitting model to feature number 57, PWY.6703..preQ0.biosynthesis
## 2024-07-16 13:51:29.393605 INFO::Fitting model to feature number 58, THISYN.PWY..superpathway.of.thiamin.diphosphate.biosynthesis.I
## 2024-07-16 13:51:29.395907 INFO::Fitting model to feature number 59, HISDEG.PWY..L.histidine.degradation.I
## 2024-07-16 13:51:29.398177 INFO::Fitting model to feature number 60, PWY.5659..GDP.mannose.biosynthesis
## 2024-07-16 13:51:29.400476 INFO::Fitting model to feature number 61, PWY.5030..L.histidine.degradation.III
## 2024-07-16 13:51:29.402735 INFO::Fitting model to feature number 62, PWY.5484..glycolysis.II..from.fructose.6.phosphate.
## 2024-07-16 13:51:29.405028 INFO::Fitting model to feature number 63, PWY.3001..superpathway.of.L.isoleucine.biosynthesis.I
## 2024-07-16 13:51:29.407331 INFO::Fitting model to feature number 64, PWY.6125..superpathway.of.guanosine.nucleotides.de.novo.biosynthesis.II
## 2024-07-16 13:51:29.409652 INFO::Fitting model to feature number 65, TRPSYN.PWY..L.tryptophan.biosynthesis
## 2024-07-16 13:51:29.411958 INFO::Fitting model to feature number 66, GLYCOLYSIS..glycolysis.I..from.glucose.6.phosphate.
## 2024-07-16 13:51:29.41423 INFO::Fitting model to feature number 67, PWY.724..superpathway.of.L.lysine..L.threonine.and.L.methionine.biosynthesis.II
## 2024-07-16 13:51:29.416522 INFO::Fitting model to feature number 68, PWY.6897..thiamin.salvage.II
## 2024-07-16 13:51:29.418814 INFO::Fitting model to feature number 69, BRANCHED.CHAIN.AA.SYN.PWY..superpathway.of.branched.amino.acid.biosynthesis
## 2024-07-16 13:51:29.42112 INFO::Fitting model to feature number 70, CALVIN.PWY..Calvin.Benson.Bassham.cycle
## 2024-07-16 13:51:29.423386 INFO::Fitting model to feature number 71, PWY.841..superpathway.of.purine.nucleotides.de.novo.biosynthesis.I
## 2024-07-16 13:51:29.425677 INFO::Fitting model to feature number 72, COMPLETE.ARO.PWY..superpathway.of.aromatic.amino.acid.biosynthesis
## 2024-07-16 13:51:29.427971 INFO::Fitting model to feature number 73, PWY.7220..adenosine.deoxyribonucleotides.de.novo.biosynthesis.II
## 2024-07-16 13:51:29.430252 INFO::Fitting model to feature number 74, PWY.7222..guanosine.deoxyribonucleotides.de.novo.biosynthesis.II
## 2024-07-16 13:51:29.432573 INFO::Fitting model to feature number 75, DENOVOPURINE2.PWY..superpathway.of.purine.nucleotides.de.novo.biosynthesis.II
## 2024-07-16 13:51:29.434907 INFO::Fitting model to feature number 76, PWY.7357..thiamin.formation.from.pyrithiamine.and.oxythiamine..yeast.
## 2024-07-16 13:51:29.437204 INFO::Fitting model to feature number 77, PWY.5103..L.isoleucine.biosynthesis.III
## 2024-07-16 13:51:29.439545 INFO::Fitting model to feature number 78, ARO.PWY..chorismate.biosynthesis.I
## 2024-07-16 13:51:29.441805 INFO::Fitting model to feature number 79, PWY.7282..4.amino.2.methyl.5.phosphomethylpyrimidine.biosynthesis..yeast.
## 2024-07-16 13:51:29.444149 INFO::Fitting model to feature number 80, PWY0.845..superpathway.of.pyridoxal.5..phosphate.biosynthesis.and.salvage
## 2024-07-16 13:51:29.446412 INFO::Fitting model to feature number 81, ANAEROFRUCAT.PWY..homolactic.fermentation
## 2024-07-16 13:51:29.4487 INFO::Fitting model to feature number 82, PWY.6892..thiazole.biosynthesis.I..E..coli.
## 2024-07-16 13:51:29.45095 INFO::Fitting model to feature number 83, RHAMCAT.PWY..L.rhamnose.degradation.I
## 2024-07-16 13:51:29.45328 INFO::Fitting model to feature number 84, PYRIDOXSYN.PWY..pyridoxal.5..phosphate.biosynthesis.I
## 2024-07-16 13:51:29.455552 INFO::Fitting model to feature number 85, PWY.1269..CMP.3.deoxy.D.manno.octulosonate.biosynthesis.I
## 2024-07-16 13:51:29.457822 INFO::Fitting model to feature number 86, PHOSLIPSYN.PWY..superpathway.of.phospholipid.biosynthesis.I..bacteria.
## 2024-07-16 13:51:29.460135 INFO::Fitting model to feature number 87, CITRULBIO.PWY..L.citrulline.biosynthesis
## 2024-07-16 13:51:29.462516 INFO::Fitting model to feature number 88, NAGLIPASYN.PWY..lipid.IVA.biosynthesis
## 2024-07-16 13:51:29.4648 INFO::Fitting model to feature number 89, BIOTIN.BIOSYNTHESIS.PWY..biotin.biosynthesis.I
## 2024-07-16 13:51:29.467057 INFO::Fitting model to feature number 90, PWY.5154..L.arginine.biosynthesis.III..via.N.acetyl.L.citrulline.
## 2024-07-16 13:51:29.469365 INFO::Fitting model to feature number 91, FASYN.ELONG.PWY..fatty.acid.elongation....saturated
## 2024-07-16 13:51:29.471647 INFO::Fitting model to feature number 92, PENTOSE.P.PWY..pentose.phosphate.pathway
## 2024-07-16 13:51:29.473919 INFO::Fitting model to feature number 93, PWYG.321..mycolate.biosynthesis
## 2024-07-16 13:51:29.47625 INFO::Fitting model to feature number 94, PWY.6519..8.amino.7.oxononanoate.biosynthesis.I
## 2024-07-16 13:51:29.478527 INFO::Fitting model to feature number 95, PWY.7664..oleate.biosynthesis.IV..anaerobic.
## 2024-07-16 13:51:29.48082 INFO::Fitting model to feature number 96, PWY.4981..L.proline.biosynthesis.II..from.arginine.
## 2024-07-16 13:51:29.483078 INFO::Fitting model to feature number 97, GLYCOGENSYNTH.PWY..glycogen.biosynthesis.I..from.ADP.D.Glucose.
## 2024-07-16 13:51:29.485417 INFO::Fitting model to feature number 98, PWY.7388..octanoyl..acyl.carrier.protein..biosynthesis..mitochondria..yeast.
## 2024-07-16 13:51:29.487756 INFO::Fitting model to feature number 99, NONOXIPENT.PWY..pentose.phosphate.pathway..non.oxidative.branch.
## 2024-07-16 13:51:29.490022 INFO::Fitting model to feature number 100, COBALSYN.PWY..adenosylcobalamin.salvage.from.cobinamide.I
## 2024-07-16 13:51:29.492338 INFO::Fitting model to feature number 101, PWY.5989..stearate.biosynthesis.II..bacteria.and.plants.
## 2024-07-16 13:51:29.494623 INFO::Fitting model to feature number 102, PWY.6282..palmitoleate.biosynthesis.I..from..5Z..dodec.5.enoate.
## 2024-07-16 13:51:29.496914 INFO::Fitting model to feature number 103, PWY0.862...5Z..dodec.5.enoate.biosynthesis
## 2024-07-16 13:51:29.499176 INFO::Fitting model to feature number 104, PWY.7400..L.arginine.biosynthesis.IV..archaebacteria.
## 2024-07-16 13:51:29.501496 INFO::Fitting model to feature number 105, ARGSYN.PWY..L.arginine.biosynthesis.I..via.L.ornithine.
## 2024-07-16 13:51:29.503832 INFO::Fitting model to feature number 106, PWY.4984..urea.cycle
## 2024-07-16 13:51:29.506102 INFO::Fitting model to feature number 107, FASYN.INITIAL.PWY..superpathway.of.fatty.acid.biosynthesis.initiation..E..coli.
## 2024-07-16 13:51:29.508378 INFO::Fitting model to feature number 108, PWY4FS.7..phosphatidylglycerol.biosynthesis.I..plastidic.
## 2024-07-16 13:51:29.510629 INFO::Fitting model to feature number 109, PWY4FS.8..phosphatidylglycerol.biosynthesis.II..non.plastidic.
## 2024-07-16 13:51:29.512948 INFO::Fitting model to feature number 110, ARGSYNBSUB.PWY..L.arginine.biosynthesis.II..acetyl.cycle.
## 2024-07-16 13:51:29.515212 INFO::Fitting model to feature number 111, PWY.6317..galactose.degradation.I..Leloir.pathway.
## 2024-07-16 13:51:29.517498 INFO::Fitting model to feature number 112, PWY66.422..D.galactose.degradation.V..Leloir.pathway.
## 2024-07-16 13:51:29.519799 INFO::Fitting model to feature number 113, GLUTORN.PWY..L.ornithine.biosynthesis
## 2024-07-16 13:51:29.522051 INFO::Fitting model to feature number 114, PWY.2941..L.lysine.biosynthesis.II
## 2024-07-16 13:51:29.524342 INFO::Fitting model to feature number 115, PWY0.1296..purine.ribonucleosides.degradation
## 2024-07-16 13:51:29.526596 INFO::Fitting model to feature number 116, NAD.BIOSYNTHESIS.II..NAD.salvage.pathway.II
## 2024-07-16 13:51:29.528911 INFO::Fitting model to feature number 117, HSERMETANA.PWY..L.methionine.biosynthesis.III
## 2024-07-16 13:51:29.531176 INFO::Fitting model to feature number 118, PWY0.162..superpathway.of.pyrimidine.ribonucleotides.de.novo.biosynthesis
## 2024-07-16 13:51:29.533485 INFO::Fitting model to feature number 119, PWY.7208..superpathway.of.pyrimidine.nucleobases.salvage
## 2024-07-16 13:51:29.535873 INFO::Fitting model to feature number 120, TEICHOICACID.PWY..teichoic.acid..poly.glycerol..biosynthesis
## 2024-07-16 13:51:29.538252 INFO::Fitting model to feature number 121, PWY.6305..putrescine.biosynthesis.IV
## 2024-07-16 13:51:29.540593 INFO::Fitting model to feature number 122, GLUCONEO.PWY..gluconeogenesis.I
## 2024-07-16 13:51:29.542871 INFO::Fitting model to feature number 123, PWY.621..sucrose.degradation.III..sucrose.invertase.
## 2024-07-16 13:51:29.545205 INFO::Fitting model to feature number 124, PWY.6901..superpathway.of.glucose.and.xylose.degradation
## 2024-07-16 13:51:29.547543 INFO::Fitting model to feature number 125, POLYISOPRENSYN.PWY..polyisoprenoid.biosynthesis..E..coli.
## 2024-07-16 13:51:29.549845 INFO::Fitting model to feature number 126, PWY.7323..superpathway.of.GDP.mannose.derived.O.antigen.building.blocks.biosynthesis
## 2024-07-16 13:51:29.552204 INFO::Fitting model to feature number 127, PWY.5941..glycogen.degradation.II..eukaryotic.
## 2024-07-16 13:51:29.554451 INFO::Fitting model to feature number 128, PWY.6527..stachyose.degradation
## 2024-07-16 13:51:29.556773 INFO::Fitting model to feature number 129, PWY0.1261..anhydromuropeptides.recycling
## 2024-07-16 13:51:29.559041 INFO::Fitting model to feature number 130, PWY66.400..glycolysis.VI..metazoan.
## 2024-07-16 13:51:29.561355 INFO::Fitting model to feature number 131, PWY0.166..superpathway.of.pyrimidine.deoxyribonucleotides.de.novo.biosynthesis..E..coli.
## 2024-07-16 13:51:29.563705 INFO::Fitting model to feature number 132, GLYCOLYSIS.E.D..superpathway.of.glycolysis.and.Entner.Doudoroff
## 2024-07-16 13:51:29.565984 INFO::Fitting model to feature number 133, PWY.7187..pyrimidine.deoxyribonucleotides.de.novo.biosynthesis.II
## 2024-07-16 13:51:29.568314 INFO::Fitting model to feature number 134, PWY.7456..mannan.degradation
## 2024-07-16 13:51:29.570595 INFO::Fitting model to feature number 135, PWY.7184..pyrimidine.deoxyribonucleotides.de.novo.biosynthesis.I
## 2024-07-16 13:51:29.5729 INFO::Fitting model to feature number 136, OANTIGEN.PWY..O.antigen.building.blocks.biosynthesis..E..coli.
## 2024-07-16 13:51:29.575178 INFO::Fitting model to feature number 137, COLANSYN.PWY..colanic.acid.building.blocks.biosynthesis
## 2024-07-16 13:51:29.577513 INFO::Fitting model to feature number 138, PWY.6545..pyrimidine.deoxyribonucleotides.de.novo.biosynthesis.III
## 2024-07-16 13:51:29.579864 INFO::Fitting model to feature number 139, PWY.6895..superpathway.of.thiamin.diphosphate.biosynthesis.II
## 2024-07-16 13:51:29.582159 INFO::Fitting model to feature number 140, PWY.6608..guanosine.nucleotides.degradation.III
## 2024-07-16 13:51:29.584441 INFO::Fitting model to feature number 141, TCA..TCA.cycle.I..prokaryotic.
## 2024-07-16 13:51:29.586723 INFO::Fitting model to feature number 142, PWY.6859..all.trans.farnesol.biosynthesis
## 2024-07-16 13:51:29.58903 INFO::Fitting model to feature number 143, PWY.5100..pyruvate.fermentation.to.acetate.and.lactate.II
## 2024-07-16 13:51:29.591331 INFO::Fitting model to feature number 144, PWY.6572..chondroitin.sulfate.degradation.I..bacterial.
## 2024-07-16 13:51:29.593635 INFO::Fitting model to feature number 145, PWY.6471..peptidoglycan.biosynthesis.IV..Enterococcus.faecium.
## 2024-07-16 13:51:29.595928 INFO::Fitting model to feature number 146, PWY.6470..peptidoglycan.biosynthesis.V...beta..lactam.resistance.
## 2024-07-16 13:51:29.598183 INFO::Fitting model to feature number 147, PWY.5188..tetrapyrrole.biosynthesis.I..from.glutamate.
## 2024-07-16 13:51:29.600517 INFO::Fitting model to feature number 148, GLUCUROCAT.PWY..superpathway.of..beta..D.glucuronide.and.D.glucuronate.degradation
## 2024-07-16 13:51:29.602804 INFO::Fitting model to feature number 149, PWY.6749..CMP.legionaminate.biosynthesis.I
## 2024-07-16 13:51:29.605151 INFO::Fitting model to feature number 150, P164.PWY..purine.nucleobases.degradation.I..anaerobic.
## 2024-07-16 13:51:29.607465 INFO::Fitting model to feature number 151, PWY.7198..pyrimidine.deoxyribonucleotides.de.novo.biosynthesis.IV
## 2024-07-16 13:51:29.609802 INFO::Fitting model to feature number 152, PWY.7211..superpathway.of.pyrimidine.deoxyribonucleotides.de.novo.biosynthesis
## 2024-07-16 13:51:29.612188 INFO::Fitting model to feature number 153, PWY66.409..superpathway.of.purine.nucleotide.salvage
## 2024-07-16 13:51:29.614894 INFO::Fitting model to feature number 154, GALACT.GLUCUROCAT.PWY..superpathway.of.hexuronide.and.hexuronate.degradation
## 2024-07-16 13:51:29.618282 INFO::Fitting model to feature number 155, UDPNAGSYN.PWY..UDP.N.acetyl.D.glucosamine.biosynthesis.I
## 2024-07-16 13:51:29.621481 INFO::Fitting model to feature number 156, PWY.6353..purine.nucleotides.degradation.II..aerobic.
## 2024-07-16 13:51:29.624669 INFO::Fitting model to feature number 157, PWY.7197..pyrimidine.deoxyribonucleotide.phosphorylation
## 2024-07-16 13:51:29.62779 INFO::Fitting model to feature number 158, PWY.7242..D.fructuronate.degradation
## 2024-07-16 13:51:29.630085 INFO::Fitting model to feature number 159, PWY.7196..superpathway.of.pyrimidine.ribonucleosides.salvage
## 2024-07-16 13:51:29.632399 INFO::Fitting model to feature number 160, PWY.5690..TCA.cycle.II..plants.and.fungi.
## 2024-07-16 13:51:29.634767 INFO::Fitting model to feature number 161, PWY.5177..glutaryl.CoA.degradation
## 2024-07-16 13:51:29.6371 INFO::Fitting model to feature number 162, PWY.6478..GDP.D.glycero..alpha..D.manno.heptose.biosynthesis
## 2024-07-16 13:51:29.639395 INFO::Fitting model to feature number 163, HEXITOLDEGSUPER.PWY..superpathway.of.hexitol.degradation..bacteria.
## 2024-07-16 13:51:29.641763 INFO::Fitting model to feature number 164, GALACTUROCAT.PWY..D.galacturonate.degradation.I
## 2024-07-16 13:51:29.644069 INFO::Fitting model to feature number 165, PWY.5104..L.isoleucine.biosynthesis.IV
## 2024-07-16 13:51:29.646346 INFO::Fitting model to feature number 166, PWY.5505..L.glutamate.and.L.glutamine.biosynthesis
## 2024-07-16 13:51:29.648667 INFO::Fitting model to feature number 167, P441.PWY..superpathway.of.N.acetylneuraminate.degradation
## 2024-07-16 13:51:29.650965 INFO::Fitting model to feature number 168, PWY.6606..guanosine.nucleotides.degradation.II
## 2024-07-16 13:51:29.653288 INFO::Fitting model to feature number 169, SALVADEHYPOX.PWY..adenosine.nucleotides.degradation.II
## 2024-07-16 13:51:29.655604 INFO::Fitting model to feature number 170, PWY.5005..biotin.biosynthesis.II
## 2024-07-16 13:51:29.657904 INFO::Fitting model to feature number 171, PWY.6549..L.glutamine.biosynthesis.III
## 2024-07-16 13:51:29.660212 INFO::Fitting model to feature number 172, GLCMANNANAUT.PWY..superpathway.of.N.acetylglucosamine..N.acetylmannosamine.and.N.acetylneuraminate.degradation
## 2024-07-16 13:51:29.662499 INFO::Fitting model to feature number 173, PWY0.1241..ADP.L.glycero..beta..D.manno.heptose.biosynthesis
## 2024-07-16 13:51:29.664825 INFO::Fitting model to feature number 174, PWY.6507..4.deoxy.L.threo.hex.4.enopyranuronate.degradation
## 2024-07-16 13:51:29.667109 INFO::Fitting model to feature number 175, GALACTARDEG.PWY..D.galactarate.degradation.I
## 2024-07-16 13:51:29.669408 INFO::Fitting model to feature number 176, GLUCARGALACTSUPER.PWY..superpathway.of.D.glucarate.and.D.galactarate.degradation
## 2024-07-16 13:51:29.671711 INFO::Fitting model to feature number 177, PWY.5676..acetyl.CoA.fermentation.to.butanoate.II
## 2024-07-16 13:51:29.674017 INFO::Fitting model to feature number 178, PWY.5347..superpathway.of.L.methionine.biosynthesis..transsulfuration.
## 2024-07-16 13:51:29.676309 INFO::Fitting model to feature number 179, PWY.5138..unsaturated..even.numbered.fatty.acid..beta..oxidation
## 2024-07-16 13:51:29.678716 INFO::Fitting model to feature number 180, MET.SAM.PWY..superpathway.of.S.adenosyl.L.methionine.biosynthesis
## 2024-07-16 13:51:29.681078 INFO::Fitting model to feature number 181, PWY66.399..gluconeogenesis.III
## 2024-07-16 13:51:29.683395 INFO::Fitting model to feature number 182, PWY0.1297..superpathway.of.purine.deoxyribonucleosides.degradation
## 2024-07-16 13:51:29.685741 INFO::Fitting model to feature number 183, METSYN.PWY..L.homoserine.and.L.methionine.biosynthesis
## 2024-07-16 13:51:29.688085 INFO::Fitting model to feature number 184, PWY.7332..superpathway.of.UDP.N.acetylglucosamine.derived.O.antigen.building.blocks.biosynthesis
## 2024-07-16 13:51:29.690391 INFO::Fitting model to feature number 185, PWY.6588..pyruvate.fermentation.to.acetone
## 2024-07-16 13:51:29.692728 INFO::Fitting model to feature number 186, PWY.6891..thiazole.biosynthesis.II..Bacillus.
## 2024-07-16 13:51:29.695037 INFO::Fitting model to feature number 187, GLUCARDEG.PWY..D.glucarate.degradation.I
## 2024-07-16 13:51:29.697389 INFO::Fitting model to feature number 188, METH.ACETATE.PWY..methanogenesis.from.acetate
## 2024-07-16 13:51:29.699717 INFO::Fitting model to feature number 189, PWY.7013..L.1.2.propanediol.degradation
## 2024-07-16 13:51:29.702019 INFO::Fitting model to feature number 190, PWY.6590..superpathway.of.Clostridium.acetobutylicum.acidogenic.fermentation
## 2024-07-16 13:51:29.704379 INFO::Fitting model to feature number 191, FAO.PWY..fatty.acid..beta..oxidation.I
## 2024-07-16 13:51:29.706686 INFO::Fitting model to feature number 192, PWY.6318..L.phenylalanine.degradation.IV..mammalian..via.side.chain.
## 2024-07-16 13:51:29.709013 INFO::Fitting model to feature number 193, CENTFERM.PWY..pyruvate.fermentation.to.butanoate
## 2024-07-16 13:51:29.711321 INFO::Fitting model to feature number 194, GOLPDLCAT.PWY..superpathway.of.glycerol.degradation.to.1.3.propanediol
## 2024-07-16 13:51:29.713665 INFO::Fitting model to feature number 195, PWY.5136..fatty.acid..beta..oxidation.II..peroxisome.
## 2024-07-16 13:51:29.716004 INFO::Fitting model to feature number 196, PWY66.391..fatty.acid..beta..oxidation.VI..peroxisome.
## 2024-07-16 13:51:29.718304 INFO::Fitting model to feature number 197, HOMOSER.METSYN.PWY..L.methionine.biosynthesis.I
## 2024-07-16 13:51:29.72065 INFO::Fitting model to feature number 198, METHGLYUT.PWY..superpathway.of.methylglyoxal.degradation
## 2024-07-16 13:51:29.722959 INFO::Fitting model to feature number 199, PWY.7288..fatty.acid..beta..oxidation..peroxisome..yeast.
## 2024-07-16 13:51:29.725338 INFO::Fitting model to feature number 200, PWY.7383..anaerobic.energy.metabolism..invertebrates..cytosol.
## 2024-07-16 13:51:29.727683 INFO::Fitting model to feature number 201, P162.PWY..L.glutamate.degradation.V..via.hydroxyglutarate.
## 2024-07-16 13:51:29.73005 INFO::Fitting model to feature number 202, PWY.4041...gamma..glutamyl.cycle
## 2024-07-16 13:51:29.732435 INFO::Fitting model to feature number 203, P108.PWY..pyruvate.fermentation.to.propanoate.I
## 2024-07-16 13:51:29.734748 INFO::Fitting model to feature number 204, PWY.5022..4.aminobutanoate.degradation.V
## 2024-07-16 13:51:29.737115 INFO::Fitting model to feature number 205, GLUDEG.I.PWY..GABA.shunt
## 2024-07-16 13:51:29.739415 INFO::Fitting model to feature number 206, PWY.5677..succinate.fermentation.to.butanoate
## 2024-07-16 13:51:29.741747 INFO::Fitting model to feature number 207, P163.PWY..L.lysine.fermentation.to.acetate.and.butanoate
## 2024-07-16 13:51:29.74408 INFO::Fitting model to feature number 208, PWY.3781..aerobic.respiration.I..cytochrome.c.
## 2024-07-16 13:51:29.746367 INFO::Fitting model to feature number 209, PWY.5083..NAD.NADH.phosphorylation.and.dephosphorylation
## 2024-07-16 13:51:29.748686 INFO::Fitting model to feature number 210, DAPLYSINESYN.PWY..L.lysine.biosynthesis.I
## 2024-07-16 13:51:29.750985 INFO::Fitting model to feature number 211, P461.PWY..hexitol.fermentation.to.lactate..formate..ethanol.and.acetate
## 2024-07-16 13:51:29.753297 INFO::Fitting model to feature number 212, PWY.1861..formaldehyde.assimilation.II..RuMP.Cycle.
## 2024-07-16 13:51:29.755599 INFO::Fitting model to feature number 213, PWY.7312..dTDP.D..beta..fucofuranose.biosynthesis
## 2024-07-16 13:51:29.757906 INFO::Fitting model to feature number 214, PWY.6630..superpathway.of.L.tyrosine.biosynthesis
## 2024-07-16 13:51:29.760232 INFO::Fitting model to feature number 215, PWY.6595..superpathway.of.guanosine.nucleotides.degradation..plants.
## 2024-07-16 13:51:29.762531 INFO::Fitting model to feature number 216, ARG.POLYAMINE.SYN..superpathway.of.arginine.and.polyamine.biosynthesis
## 2024-07-16 13:51:29.764852 INFO::Fitting model to feature number 217, PWY.7003..glycerol.degradation.to.butanol
## 2024-07-16 13:51:29.767162 INFO::Fitting model to feature number 218, P185.PWY..formaldehyde.assimilation.III..dihydroxyacetone.cycle.
## 2024-07-16 13:51:29.769487 INFO::Fitting model to feature number 219, POLYAMSYN.PWY..superpathway.of.polyamine.biosynthesis.I
## 2024-07-16 13:51:29.771803 INFO::Fitting model to feature number 220, PWY0.1298..superpathway.of.pyrimidine.deoxyribonucleosides.degradation
## 2024-07-16 13:51:29.774087 INFO::Fitting model to feature number 221, LACTOSECAT.PWY..lactose.and.galactose.degradation.I
## 2024-07-16 13:51:29.776409 INFO::Fitting model to feature number 222, PWY4LZ.257..superpathway.of.fermentation..Chlamydomonas.reinhardtii.
## 2024-07-16 13:51:29.778694 INFO::Fitting model to feature number 223, P161.PWY..acetylene.degradation
## 2024-07-16 13:51:29.781005 INFO::Fitting model to feature number 224, RUMP.PWY..formaldehyde.oxidation.I
## 2024-07-16 13:51:29.783309 INFO::Fitting model to feature number 225, PPGPPMET.PWY..ppGpp.biosynthesis
## 2024-07-16 13:51:29.785623 INFO::Fitting model to feature number 226, PWY.5304..superpathway.of.sulfur.oxidation..Acidianus.ambivalens.
## 2024-07-16 13:51:29.787955 INFO::Fitting model to feature number 227, PWY.6531..mannitol.cycle
## 2024-07-16 13:51:29.79027 INFO::Fitting model to feature number 228, FUC.RHAMCAT.PWY..superpathway.of.fucose.and.rhamnose.degradation
## 2024-07-16 13:51:29.792608 INFO::Fitting model to feature number 229, PWY.7315..dTDP.N.acetylthomosamine.biosynthesis
## 2024-07-16 13:51:29.7949 INFO::Fitting model to feature number 230, FUCCAT.PWY..fucose.degradation
## 2024-07-16 13:51:29.797255 INFO::Fitting model to feature number 231, PWY.7210..pyrimidine.deoxyribonucleotides.biosynthesis.from.CTP
## 2024-07-16 13:51:29.799595 INFO::Fitting model to feature number 232, FOLSYN.PWY..superpathway.of.tetrahydrofolate.biosynthesis.and.salvage
## 2024-07-16 13:51:29.801903 INFO::Fitting model to feature number 233, PWY.6612..superpathway.of.tetrahydrofolate.biosynthesis
## 2024-07-16 13:51:29.804281 INFO::Fitting model to feature number 234, PWY0.1061..superpathway.of.L.alanine.biosynthesis
## 2024-07-16 13:51:29.806579 INFO::Fitting model to feature number 235, PWY.5189..tetrapyrrole.biosynthesis.II..from.glycine.
## 2024-07-16 13:51:29.808887 INFO::Fitting model to feature number 236, PWY.6876..isopropanol.biosynthesis
## 2024-07-16 13:51:29.811187 INFO::Fitting model to feature number 237, PWY0.781..aspartate.superpathway
## 2024-07-16 13:51:29.813532 INFO::Fitting model to feature number 238, P4.PWY..superpathway.of.L.lysine..L.threonine.and.L.methionine.biosynthesis.I
## 2024-07-16 13:51:29.815843 INFO::Fitting model to feature number 239, PWY0.1479..tRNA.processing
## 2024-07-16 13:51:29.818135 INFO::Fitting model to feature number 240, GLYCOLYSIS.TCA.GLYOX.BYPASS..superpathway.of.glycolysis..pyruvate.dehydrogenase..TCA..and.glyoxylate.bypass
## 2024-07-16 13:51:29.820458 INFO::Fitting model to feature number 241, PYRIDNUCSAL.PWY..NAD.salvage.pathway.I
## 2024-07-16 13:51:29.822776 INFO::Fitting model to feature number 242, GLYCOCAT.PWY..glycogen.degradation.I..bacterial.
## 2024-07-16 13:51:29.825131 INFO::Fitting model to feature number 243, PWY.5723..Rubisco.shunt
## 2024-07-16 13:51:29.827417 INFO::Fitting model to feature number 244, TCA.GLYOX.BYPASS..superpathway.of.glyoxylate.bypass.and.TCA
## 2024-07-16 13:51:29.829777 INFO::Fitting model to feature number 245, PWY.7385..1.3.propanediol.biosynthesis..engineered.
## 2024-07-16 13:51:29.832181 INFO::Fitting model to feature number 246, PWY.7254..TCA.cycle.VII..acetate.producers.
## 2024-07-16 13:51:29.834496 INFO::Fitting model to feature number 247, GLYOXYLATE.BYPASS..glyoxylate.cycle
## 2024-07-16 13:51:29.836954 INFO::Fitting model to feature number 248, PWY.6731..starch.degradation.III
## 2024-07-16 13:51:29.839246 INFO::Fitting model to feature number 249, PWY.7328..superpathway.of.UDP.glucose.derived.O.antigen.building.blocks.biosynthesis
## 2024-07-16 13:51:29.841608 INFO::Fitting model to feature number 250, POLYAMINSYN3.PWY..superpathway.of.polyamine.biosynthesis.II
## 2024-07-16 13:51:29.843963 INFO::Fitting model to feature number 251, PWY.2723..trehalose.degradation.V
## 2024-07-16 13:51:29.846243 INFO::Fitting model to feature number 252, GLUCOSE1PMETAB.PWY..glucose.and.glucose.1.phosphate.degradation
## 2024-07-16 13:51:29.848586 INFO::Fitting model to feature number 253, PWY.5384..sucrose.degradation.IV..sucrose.phosphorylase.
## 2024-07-16 13:51:29.850868 INFO::Fitting model to feature number 254, PWY.7046..4.coumarate.degradation..anaerobic.
## 2024-07-16 13:51:29.8532 INFO::Fitting model to feature number 255, PWY66.389..phytol.degradation
## 2024-07-16 13:51:29.855515 INFO::Fitting model to feature number 256, PWY.5971..palmitate.biosynthesis.II..bacteria.and.plants.
## 2024-07-16 13:51:29.857811 INFO::Fitting model to feature number 257, PWY.6629..superpathway.of.L.tryptophan.biosynthesis
## 2024-07-16 13:51:29.860159 INFO::Fitting model to feature number 258, PWY.5345..superpathway.of.L.methionine.biosynthesis..by.sulfhydrylation.
## 2024-07-16 13:51:29.862452 INFO::Fitting model to feature number 259, SULFATE.CYS.PWY..superpathway.of.sulfate.assimilation.and.cysteine.biosynthesis
## 2024-07-16 13:51:29.864761 INFO::Fitting model to feature number 260, PWY.5838..superpathway.of.menaquinol.8.biosynthesis.I
## 2024-07-16 13:51:29.867049 INFO::Fitting model to feature number 261, PRPP.PWY..superpathway.of.histidine..purine..and.pyrimidine.biosynthesis
## 2024-07-16 13:51:29.869354 INFO::Fitting model to feature number 262, PWY.5845..superpathway.of.menaquinol.9.biosynthesis
## 2024-07-16 13:51:29.871669 INFO::Fitting model to feature number 263, PWY.6113..superpathway.of.mycolate.biosynthesis
## 2024-07-16 13:51:29.873955 INFO::Fitting model to feature number 264, PWY.5897..superpathway.of.menaquinol.11.biosynthesis
## 2024-07-16 13:51:29.876281 INFO::Fitting model to feature number 265, PWY.5898..superpathway.of.menaquinol.12.biosynthesis
## 2024-07-16 13:51:29.878583 INFO::Fitting model to feature number 266, PWY.5899..superpathway.of.menaquinol.13.biosynthesis
## 2024-07-16 13:51:29.880911 INFO::Fitting model to feature number 267, PWY.7117..C4.photosynthetic.carbon.assimilation.cycle..PEPCK.type
## 2024-07-16 13:51:29.883202 INFO::Fitting model to feature number 268, PWY.6969..TCA.cycle.V..2.oxoglutarate.ferredoxin.oxidoreductase.
## 2024-07-16 13:51:29.88552 INFO::Fitting model to feature number 269, PWY.6628..superpathway.of.L.phenylalanine.biosynthesis
## 2024-07-16 13:51:29.887833 INFO::Fitting model to feature number 270, PWY.5861..superpathway.of.demethylmenaquinol.8.biosynthesis
## 2024-07-16 13:51:29.890117 INFO::Fitting model to feature number 271, PWY.5862..superpathway.of.demethylmenaquinol.9.biosynthesis
## 2024-07-16 13:51:29.892454 INFO::Fitting model to feature number 272, FERMENTATION.PWY..mixed.acid.fermentation
## 2024-07-16 13:51:29.894755 INFO::Fitting model to feature number 273, PWY.241..C4.photosynthetic.carbon.assimilation.cycle..NADP.ME.type
## 2024-07-16 13:51:29.89711 INFO::Fitting model to feature number 274, SO4ASSIM.PWY..sulfate.reduction.I..assimilatory.
## 2024-07-16 13:51:29.899411 INFO::Fitting model to feature number 275, KETOGLUCONMET.PWY..ketogluconate.metabolism
## 2024-07-16 13:51:29.901733 INFO::Fitting model to feature number 276, PWY.5840..superpathway.of.menaquinol.7.biosynthesis
## 2024-07-16 13:51:29.904053 INFO::Fitting model to feature number 277, PWY.5850..superpathway.of.menaquinol.6.biosynthesis.I
## 2024-07-16 13:51:29.906353 INFO::Fitting model to feature number 278, PWY.5896..superpathway.of.menaquinol.10.biosynthesis
## 2024-07-16 13:51:29.908728 INFO::Fitting model to feature number 279, P105.PWY..TCA.cycle.IV..2.oxoglutarate.decarboxylase.
## 2024-07-16 13:51:29.911023 INFO::Fitting model to feature number 280, PWY.5913..TCA.cycle.VI..obligate.autotrophs.
## 2024-07-16 13:51:29.913333 INFO::Fitting model to feature number 281, PWY.561..superpathway.of.glyoxylate.cycle.and.fatty.acid.degradation
## 2024-07-16 13:51:29.915652 INFO::Fitting model to feature number 282, PWY.4702..phytate.degradation.I
## 2024-07-16 13:51:29.917941 INFO::Fitting model to feature number 283, PWY.6803..phosphatidylcholine.acyl.editing
## 2024-07-16 13:51:29.920277 INFO::Fitting model to feature number 284, PWY.7115..C4.photosynthetic.carbon.assimilation.cycle..NAD.ME.type
## 2024-07-16 13:51:29.922584 INFO::Fitting model to feature number 285, PWY.5918..superpathay.of.heme.biosynthesis.from.glutamate
## 2024-07-16 13:51:29.92491 INFO::Fitting model to feature number 286, PWY.7234..inosine.5..phosphate.biosynthesis.III
## 2024-07-16 13:51:29.927196 INFO::Fitting model to feature number 287, PWY.6285..superpathway.of.fatty.acids.biosynthesis..E..coli.
## 2024-07-16 13:51:29.929511 INFO::Fitting model to feature number 288, PWY.5860..superpathway.of.demethylmenaquinol.6.biosynthesis.I
## 2024-07-16 13:51:29.931839 INFO::Fitting model to feature number 289, PWY.5863..superpathway.of.phylloquinol.biosynthesis
## 2024-07-16 13:51:29.93416 INFO::Fitting model to feature number 290, HEME.BIOSYNTHESIS.II..heme.biosynthesis.I..aerobic.
## 2024-07-16 13:51:29.936486 INFO::Fitting model to feature number 291, PWY.821..superpathway.of.sulfur.amino.acid.biosynthesis..Saccharomyces.cerevisiae.
## 2024-07-16 13:51:29.938802 INFO::Fitting model to feature number 292, PWY.5675..nitrate.reduction.V..assimilatory.
## 2024-07-16 13:51:29.941154 INFO::Fitting model to feature number 293, PWY.5791..1.4.dihydroxy.2.naphthoate.biosynthesis.II..plants.
## 2024-07-16 13:51:29.943446 INFO::Fitting model to feature number 294, PWY.5837..1.4.dihydroxy.2.naphthoate.biosynthesis.I
## 2024-07-16 13:51:29.945769 INFO::Fitting model to feature number 295, PWY.5920..superpathway.of.heme.biosynthesis.from.glycine
## 2024-07-16 13:51:29.948101 INFO::Fitting model to feature number 296, PWY.7204..pyridoxal.5..phosphate.salvage.II..plants.
## 2024-07-16 13:51:29.976976 INFO::Fitting model to feature number 297, ENTBACSYN.PWY..enterobactin.biosynthesis
## 2024-07-16 13:51:29.981294 INFO::Fitting model to feature number 298, PWY0.1415..superpathway.of.heme.biosynthesis.from.uroporphyrinogen.III
## 2024-07-16 13:51:29.983765 INFO::Fitting model to feature number 299, HEMESYN2.PWY..heme.biosynthesis.II..anaerobic.
## 2024-07-16 13:51:29.98605 INFO::Fitting model to feature number 300, PWY.6284..superpathway.of.unsaturated.fatty.acids.biosynthesis..E..coli.
## 2024-07-16 13:51:29.988329 INFO::Fitting model to feature number 301, P42.PWY..incomplete.reductive.TCA.cycle
## 2024-07-16 13:51:29.99057 INFO::Fitting model to feature number 302, PWY.5656..mannosylglycerate.biosynthesis.I
## 2024-07-16 13:51:29.992853 INFO::Fitting model to feature number 303, PWY.5173..superpathway.of.acetyl.CoA.biosynthesis
## 2024-07-16 13:51:29.995116 INFO::Fitting model to feature number 304, ECASYN.PWY..enterobacterial.common.antigen.biosynthesis
## 2024-07-16 13:51:29.997415 INFO::Fitting model to feature number 305, AST.PWY..L.arginine.degradation.II..AST.pathway.
## 2024-07-16 13:51:29.999732 INFO::Fitting model to feature number 306, PWY3O.355..stearate.biosynthesis.III..fungi.
## 2024-07-16 13:51:30.001993 INFO::Fitting model to feature number 307, PWY0.42..2.methylcitrate.cycle.I
## 2024-07-16 13:51:30.004265 INFO::Fitting model to feature number 308, PWY.7094..fatty.acid.salvage
## 2024-07-16 13:51:30.006516 INFO::Fitting model to feature number 309, PWY.5747..2.methylcitrate.cycle.II
## 2024-07-16 13:51:30.008822 INFO::Fitting model to feature number 310, PWY.5367..petroselinate.biosynthesis
## 2024-07-16 13:51:30.011094 INFO::Fitting model to feature number 311, PWY0.1533..methylphosphonate.degradation.I
## 2024-07-16 13:51:30.013421 INFO::Fitting model to feature number 312, PWY.6823..molybdenum.cofactor.biosynthesis
## 2024-07-16 13:51:30.015729 INFO::Fitting model to feature number 313, PWY.5705..allantoin.degradation.to.glyoxylate.III
## 2024-07-16 13:51:30.017996 INFO::Fitting model to feature number 314, LPSSYN.PWY..superpathway.of.lipopolysaccharide.biosynthesis
## 2024-07-16 13:51:30.020283 INFO::Fitting model to feature number 315, REDCITCYC..TCA.cycle.VIII..helicobacter.
## 2024-07-16 13:51:30.022546 INFO::Fitting model to feature number 316, GLYCOL.GLYOXDEG.PWY..superpathway.of.glycol.metabolism.and.degradation
## 2024-07-16 13:51:30.024838 INFO::Fitting model to feature number 317, P122.PWY..heterolactic.fermentation
## 2024-07-16 13:51:30.027107 INFO::Fitting model to feature number 318, KDO.NAGLIPASYN.PWY..superpathway.of..Kdo.2.lipid.A.biosynthesis
## 2024-07-16 13:51:30.029427 INFO::Fitting model to feature number 319, PWY.7446..sulfoglycolysis
## 2024-07-16 13:51:30.031732 INFO::Fitting model to feature number 320, PWY490.3..nitrate.reduction.VI..assimilatory.
## 2024-07-16 13:51:30.033998 INFO::Fitting model to feature number 321, PWY.7209..superpathway.of.pyrimidine.ribonucleosides.degradation
## 2024-07-16 13:51:30.036426 INFO::Fitting model to feature number 322, UBISYN.PWY..superpathway.of.ubiquinol.8.biosynthesis..prokaryotic.
## 2024-07-16 13:51:30.038688 INFO::Fitting model to feature number 323, PWY0.41..allantoin.degradation.IV..anaerobic.
## 2024-07-16 13:51:30.040978 INFO::Fitting model to feature number 324, PWY.5855..ubiquinol.7.biosynthesis..prokaryotic.
## 2024-07-16 13:51:30.043264 INFO::Fitting model to feature number 325, PWY.5856..ubiquinol.9.biosynthesis..prokaryotic.
## 2024-07-16 13:51:30.045562 INFO::Fitting model to feature number 326, PWY.5857..ubiquinol.10.biosynthesis..prokaryotic.
## 2024-07-16 13:51:30.047895 INFO::Fitting model to feature number 327, PWY.6708..ubiquinol.8.biosynthesis..prokaryotic.
## 2024-07-16 13:51:30.050171 INFO::Fitting model to feature number 328, PWY.5692..allantoin.degradation.to.glyoxylate.II
## 2024-07-16 13:51:30.052479 INFO::Fitting model to feature number 329, URDEGR.PWY..superpathway.of.allantoin.degradation.in.plants
## 2024-07-16 13:51:30.054743 INFO::Fitting model to feature number 330, PWY0.1338..polymyxin.resistance
## 2024-07-16 13:51:30.057039 INFO::Fitting model to feature number 331, P221.PWY..octane.oxidation
## 2024-07-16 13:51:30.059325 INFO::Fitting model to feature number 332, THREOCAT.PWY..superpathway.of.L.threonine.metabolism
## 2024-07-16 13:51:30.061622 INFO::Fitting model to feature number 333, NONMEVIPP.PWY..methylerythritol.phosphate.pathway.I
## 2024-07-16 13:51:30.063965 INFO::Fitting model to feature number 334, PWY.7560..methylerythritol.phosphate.pathway.II
## 2024-07-16 13:51:30.066267 INFO::Fitting model to feature number 335, PWY.5121..superpathway.of.geranylgeranyl.diphosphate.biosynthesis.II..via.MEP.
## 2024-07-16 13:51:30.068573 INFO::Fitting model to feature number 336, PWY.7392..taxadiene.biosynthesis..engineered.
## 2024-07-16 13:51:30.070837 INFO::Fitting model to feature number 337, PWY.6143..CMP.pseudaminate.biosynthesis
## 2024-07-16 13:51:30.073142 INFO::Fitting model to feature number 338, PWY.6270..isoprene.biosynthesis.I
## 2024-07-16 13:51:30.075406 INFO::Fitting model to feature number 339, ALLANTOINDEG.PWY..superpathway.of.allantoin.degradation.in.yeast
## 2024-07-16 13:51:30.077798 INFO::Fitting model to feature number 340, PWY.6263..superpathway.of.menaquinol.8.biosynthesis.II
## 2024-07-16 13:51:30.080136 INFO::Fitting model to feature number 341, PWY.7371..1.4.dihydroxy.6.naphthoate.biosynthesis.II
## 2024-07-16 13:51:30.08242 INFO::Fitting model to feature number 342, ARGORNPROST.PWY..arginine..ornithine.and.proline.interconversion
## 2024-07-16 13:51:30.084723 INFO::Fitting model to feature number 343, PWY66.398..TCA.cycle.III..animals.
## 2024-07-16 13:51:30.087 INFO::Fitting model to feature number 344, PWY.5464..superpathway.of.cytosolic.glycolysis..plants...pyruvate.dehydrogenase.and.TCA.cycle
## 2024-07-16 13:51:30.089318 INFO::Fitting model to feature number 345, ORNDEG.PWY..superpathway.of.ornithine.degradation
## 2024-07-16 13:51:30.091658 INFO::Fitting model to feature number 346, P23.PWY..reductive.TCA.cycle.I
## 2024-07-16 13:51:30.093957 INFO::Fitting model to feature number 347, PWY0.1277..3.phenylpropanoate.and.3..3.hydroxyphenyl.propanoate.degradation
## 2024-07-16 13:51:30.096267 INFO::Fitting model to feature number 348, ARGDEG.PWY..superpathway.of.L.arginine..putrescine..and.4.aminobutanoate.degradation
## 2024-07-16 13:51:30.098531 INFO::Fitting model to feature number 349, ORNARGDEG.PWY..superpathway.of.L.arginine.and.L.ornithine.degradation
## 2024-07-16 13:51:30.100836 INFO::Fitting model to feature number 350, HCAMHPDEG.PWY..3.phenylpropanoate.and.3..3.hydroxyphenyl.propanoate.degradation.to.2.oxopent.4.enoate
## 2024-07-16 13:51:30.103143 INFO::Fitting model to feature number 351, PWY.6690..cinnamate.and.3.hydroxycinnamate.degradation.to.2.oxopent.4.enoate
## 2024-07-16 13:51:30.105427 INFO::Fitting model to feature number 352, PROTOCATECHUATE.ORTHO.CLEAVAGE.PWY..protocatechuate.degradation.II..ortho.cleavage.pathway.
## 2024-07-16 13:51:30.107708 INFO::Fitting model to feature number 353, P124.PWY..Bifidobacterium.shunt
## 2024-07-16 13:51:30.109997 INFO::Fitting model to feature number 354, PWY.622..starch.biosynthesis
## 2024-07-16 13:51:30.112318 INFO::Fitting model to feature number 355, PWY.7269..NAD.NADP.NADH.NADPH.mitochondrial.interconversion..yeast.
## 2024-07-16 13:51:30.114584 INFO::Fitting model to feature number 356, PWY.7316..dTDP.N.acetylviosamine.biosynthesis
## 2024-07-16 13:51:30.116924 INFO::Fitting model to feature number 357, PWY.7279..aerobic.respiration.II..cytochrome.c...yeast.
## 2024-07-16 13:51:30.119186 INFO::Fitting model to feature number 358, PWY.6837..fatty.acid.beta.oxidation.V..unsaturated..odd.number..di.isomerase.dependent.
## 2024-07-16 13:51:30.121513 INFO::Fitting model to feature number 359, X3.HYDROXYPHENYLACETATE.DEGRADATION.PWY..4.hydroxyphenylacetate.degradation
## 2024-07-16 13:51:30.123824 INFO::Fitting model to feature number 360, PWY.6953..dTDP.3.acetamido.3.6.dideoxy..alpha..D.galactose.biosynthesis
## 2024-07-16 13:51:30.12611 INFO::Fitting model to feature number 361, PWY.7616..methanol.oxidation.to.carbon.dioxide
## 2024-07-16 13:51:30.128395 INFO::Fitting model to feature number 362, PWY.5088..L.glutamate.degradation.VIII..to.propanoate.
## 2024-07-16 13:51:30.130646 INFO::Fitting model to feature number 363, PWY0.881..superpathway.of.fatty.acid.biosynthesis.I..E..coli.
## 2024-07-16 13:51:30.179374 INFO::Counting total values for each feature
## 2024-07-16 13:51:30.209945 INFO::Writing filtered data to file output_pwys/features/filtered_data.tsv
## 2024-07-16 13:51:30.2486 INFO::Writing filtered, normalized data to file output_pwys/features/filtered_data_norm.tsv
## 2024-07-16 13:51:30.286653 INFO::Writing filtered, normalized, transformed data to file output_pwys/features/filtered_data_norm_transformed.tsv
## 2024-07-16 13:51:30.336093 INFO::Writing residuals to file output_pwys/fits/residuals.rds
## 2024-07-16 13:51:30.359458 INFO::Writing fitted values to file output_pwys/fits/fitted.rds
## 2024-07-16 13:51:30.375632 INFO::Writing all results to file (ordered by increasing q-values): output_pwys/all_results.tsv
## 2024-07-16 13:51:30.381774 INFO::Writing the significant results (those which are less than or equal to the threshold of 0.250000 ) to file (ordered by increasing q-values): output_pwys/significant_results.tsv
## 2024-07-16 13:51:30.382522 INFO::Writing heatmap of significant results to file: output_pwys/heatmap.pdf
## [1] "There are no associations to plot!"
## 2024-07-16 13:51:30.383451 INFO::Writing association plots (one for each significant association) to output folder: output_pwys
## 2024-07-16 13:51:30.386709 INFO::Plotting associations from most to least significant, grouped by metadata
## 2024-07-16 13:51:30.387249 INFO::Plotting data for metadata number 1, antibiotics_current_use
## 2024-07-16 13:51:30.388316 INFO::Creating boxplot for categorical data, antibiotics_current_use vs POLYAMINSYN3.PWY..superpathway.of.polyamine.biosynthesis.II
##########################
# Extract the residuals #
##########################
datExpr <- as.data.frame(t(fit_data$residuals))
########################
# Create WGCNA modules #
########################
gsg = goodSamplesGenes(datExpr, verbose = 3)
## Flagging genes and samples with too many missing values...
## ..step 1
## ..Excluding 6 samples from the calculation due to too many missing genes.
## ..step 2
gsg$allOK
## [1] FALSE
if (!gsg$allOK)
{if (sum(!gsg$goodGenes) > 0)
printFlush(paste("Removing genes:", paste(names(datExpr)[!gsg$goodGenes], collapse = ", ")));
if (sum(!gsg$goodSamples) > 0)
printFlush(paste("Removing samples:", paste(rownames(datExpr)[!gsg$goodSamples], collapse =", ")))
datExpr = datExpr[gsg$goodSamples, gsg$goodGenes]
}
## Removing samples: CSM5MCVB_P, CSM79HNY_P, ESM5GEYY_P, ESM718U9_P, MSM6J2N6_P, MSM9VZLX_P
gsg = goodSamplesGenes(datExpr, verbose = 3)
## Flagging genes and samples with too many missing values...
## ..step 1
gsg$allOK # TRUE
## [1] TRUE
###################################
# Choose soft threshold parameter #
###################################
powers = c(c(1:20), seq(from = 22, to=30, by=2))
sft = pickSoftThreshold(datExpr, powerVector = powers, verbose = 5, dataIsExpr = TRUE, RsquaredCut = 0.30)
## pickSoftThreshold: will use block size 363.
## pickSoftThreshold: calculating connectivity for given powers...
## ..working on genes 1 through 363 of 363
## Power SFT.R.sq slope truncated.R.sq mean.k. median.k. max.k.
## 1 1 0.722 1.8100 0.7110 124.00 130.000 162.00
## 2 2 0.596 0.6920 0.6290 63.10 68.700 95.70
## 3 3 0.393 0.2300 0.2310 38.40 39.700 69.30
## 4 4 0.124 -0.0902 0.0162 25.90 24.800 55.00
## 5 5 0.561 -0.3550 0.5860 18.70 16.300 46.60
## 6 6 0.673 -0.5190 0.6680 14.20 11.400 40.40
## 7 7 0.781 -0.6230 0.8100 11.20 8.380 35.60
## 8 8 0.815 -0.7040 0.8450 9.07 6.520 31.80
## 9 9 0.861 -0.8180 0.9260 7.54 5.100 28.60
## 10 10 0.847 -0.8620 0.9070 6.39 4.100 26.00
## 11 11 0.835 -0.9150 0.9140 5.50 3.430 23.80
## 12 12 0.847 -0.9540 0.9130 4.80 2.870 21.90
## 13 13 0.868 -0.9730 0.9370 4.24 2.380 20.20
## 14 14 0.864 -0.9990 0.9160 3.78 2.060 18.70
## 15 15 0.890 -1.0300 0.9590 3.40 1.720 17.40
## 16 16 0.898 -1.0400 0.9670 3.09 1.530 16.20
## 17 17 0.888 -1.0600 0.9570 2.82 1.370 15.20
## 18 18 0.877 -1.0600 0.9290 2.59 1.280 14.20
## 19 19 0.864 -1.0500 0.9170 2.40 1.180 13.30
## 20 20 0.844 -1.0600 0.8690 2.23 1.100 12.50
## 21 22 0.854 -1.0700 0.8890 1.95 0.997 11.20
## 22 24 0.820 -1.0600 0.8720 1.74 0.899 9.98
## 23 26 0.797 -0.9790 0.8720 1.57 0.793 8.97
## 24 28 0.829 -0.9390 0.8720 1.43 0.718 8.10
## 25 30 0.807 -0.9130 0.8140 1.31 0.673 7.35
##############################
# One-step module detection #
##############################
power = sft$powerEstimate
net = blockwiseModules(datExpr,
power = power,
corFnc = "bicor",
corOptions = list(maxPOutliers = 0.1),
networkType ="unsigned",
maxBlockSize = ncol(datExpr),
minModuleSize = 3,
TOMType = "unsigned",
reassignThreshold = 0,
mergeCutHeight = 0,
verbose = 3)
## Calculating module eigengenes block-wise from all genes
## Flagging genes and samples with too many missing values...
## ..step 1
## ..Working on block 1 .
## TOM calculation: adjacency..
## ..will not use multithreading.
## Fraction of slow calculations: 0.000000
## ..connectivity..
## ..matrix multiplication (system BLAS)..
## ..normalization..
## ..done.
## ....clustering..
## ....detecting modules..
## ....calculating module eigengenes..
## ....checking kME in modules..
## ..removing 3 genes from module 1 because their KME is too low.
## ..removing 11 genes from module 2 because their KME is too low.
## ..removing 2 genes from module 3 because their KME is too low.
## ..merging modules that are too close..
## mergeCloseModules: Merging modules whose distance is less than 0
## Calculating new MEs...
####################
# How many modules #
####################
ncol(net$MEs)
## [1] 4
table(net$colors)
##
## blue brown grey turquoise
## 139 18 16 190
##########################
# Plot module dendrogram #
##########################
eigenGenes <- net$MEs
MEDiss = 1-cor(eigenGenes)
METree = hclust(as.dist(MEDiss), method = "average")
plot(METree, main = "Clustering of module eigengenes", xlab = "", sub = "")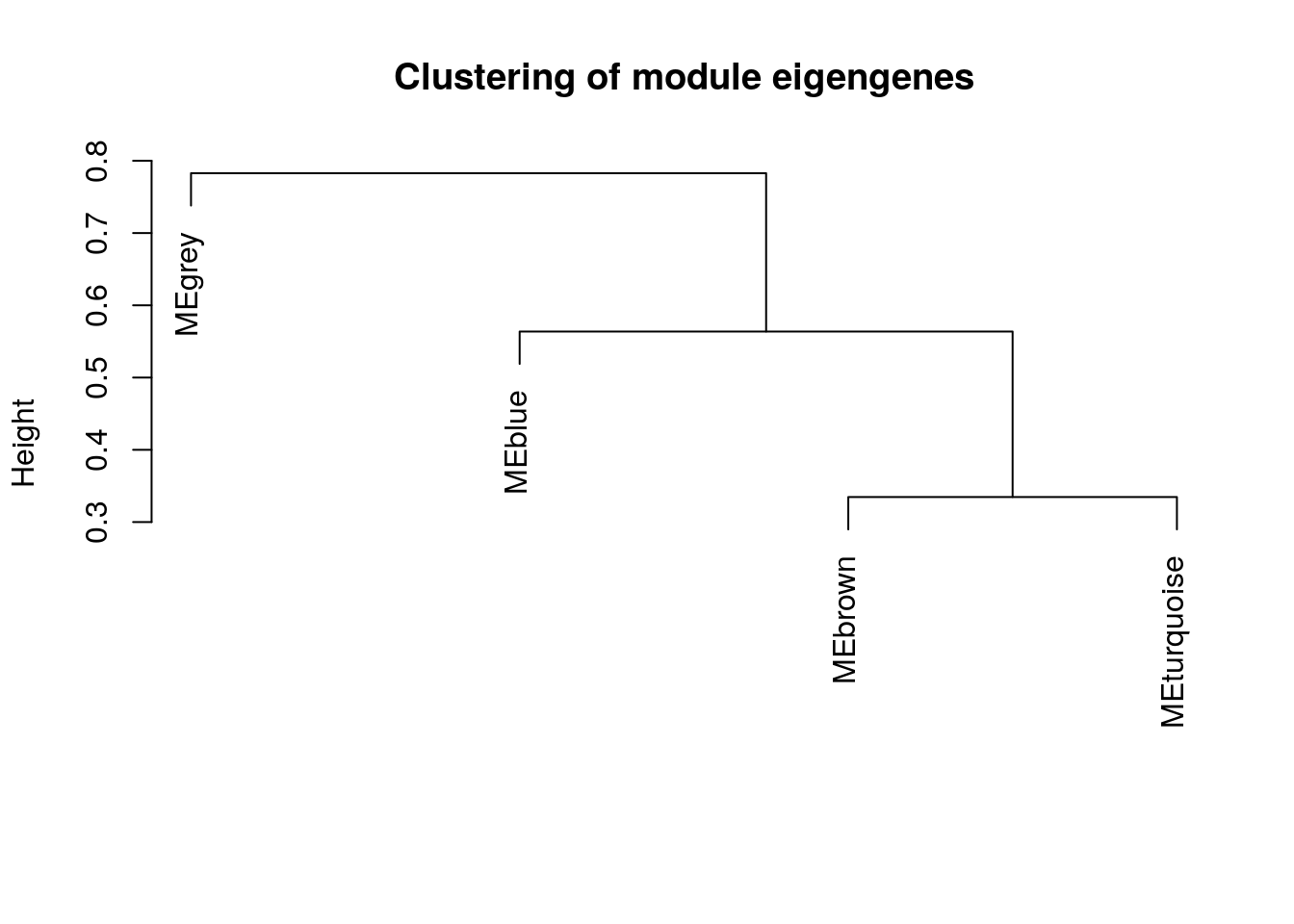
###########################################
# Re-calculate modules and find hub genes #
###########################################
moduleColors <- net$colors
MEs0 = moduleEigengenes(datExpr, moduleColors)$eigengenes
modules_data = orderMEs(MEs0)
#######################
# Create mapping file #
#######################
feature_by_modules <- as.data.frame(net$colors)
feature_by_modules <- rownames_to_column(feature_by_modules)
colnames(feature_by_modules) <- c('Feature', 'Module')
features_mapping <- feature_by_modules
features_mapping$Module <- paste('ME', features_mapping$Module, sep = '')We perform the MSEA as before using the modules from the WGCNA analysis on the pathways.
###################
# Rank DA results #
###################
results <- fit_data$results %>% filter(metadata=='disease')
results$qval <- p.adjust(results$pval, 'BH')
sum(results$qval < 0.05)
## [1] 0
results <- results[order(results$qval, decreasing = FALSE),]
###################
# MSEA Processing #
##################
module_map <- features_mapping
mod.gs <- tapply(module_map$Module, module_map$Feature, as.character)
microbeSet <- inverseList(mod.gs)
microbeSet
## $MEblue
## [1] "ANAEROFRUCAT.PWY..homolactic.fermentation"
## [2] "ANAGLYCOLYSIS.PWY..glycolysis.III..from.glucose."
## [3] "ARGININE.SYN4.PWY..L.ornithine.de.novo..biosynthesis"
## [4] "ARGSYN.PWY..L.arginine.biosynthesis.I..via.L.ornithine."
## [5] "ARGSYNBSUB.PWY..L.arginine.biosynthesis.II..acetyl.cycle."
## [6] "ARO.PWY..chorismate.biosynthesis.I"
## [7] "ASPASN.PWY..superpathway.of.L.aspartate.and.L.asparagine.biosynthesis"
## [8] "BRANCHED.CHAIN.AA.SYN.PWY..superpathway.of.branched.amino.acid.biosynthesis"
## [9] "CALVIN.PWY..Calvin.Benson.Bassham.cycle"
## [10] "CITRULBIO.PWY..L.citrulline.biosynthesis"
## [11] "COA.PWY..coenzyme.A.biosynthesis.I"
## [12] "COA.PWY.1..coenzyme.A.biosynthesis.II..mammalian."
## [13] "COBALSYN.PWY..adenosylcobalamin.salvage.from.cobinamide.I"
## [14] "COLANSYN.PWY..colanic.acid.building.blocks.biosynthesis"
## [15] "COMPLETE.ARO.PWY..superpathway.of.aromatic.amino.acid.biosynthesis"
## [16] "DENOVOPURINE2.PWY..superpathway.of.purine.nucleotides.de.novo.biosynthesis.II"
## [17] "DTDPRHAMSYN.PWY..dTDP.L.rhamnose.biosynthesis.I"
## [18] "GALACT.GLUCUROCAT.PWY..superpathway.of.hexuronide.and.hexuronate.degradation"
## [19] "GALACTUROCAT.PWY..D.galacturonate.degradation.I"
## [20] "GLUCONEO.PWY..gluconeogenesis.I"
## [21] "GLUCUROCAT.PWY..superpathway.of..beta..D.glucuronide.and.D.glucuronate.degradation"
## [22] "GLUTORN.PWY..L.ornithine.biosynthesis"
## [23] "GLYCOGENSYNTH.PWY..glycogen.biosynthesis.I..from.ADP.D.Glucose."
## [24] "GLYCOLYSIS..glycolysis.I..from.glucose.6.phosphate."
## [25] "HISDEG.PWY..L.histidine.degradation.I"
## [26] "HISTSYN.PWY..L.histidine.biosynthesis"
## [27] "HSERMETANA.PWY..L.methionine.biosynthesis.III"
## [28] "ILEUSYN.PWY..L.isoleucine.biosynthesis.I..from.threonine."
## [29] "NAGLIPASYN.PWY..lipid.IVA.biosynthesis"
## [30] "NONOXIPENT.PWY..pentose.phosphate.pathway..non.oxidative.branch."
## [31] "OANTIGEN.PWY..O.antigen.building.blocks.biosynthesis..E..coli."
## [32] "P108.PWY..pyruvate.fermentation.to.propanoate.I"
## [33] "PANTO.PWY..phosphopantothenate.biosynthesis.I"
## [34] "PANTOSYN.PWY..pantothenate.and.coenzyme.A.biosynthesis.I"
## [35] "PENTOSE.P.PWY..pentose.phosphate.pathway"
## [36] "PEPTIDOGLYCANSYN.PWY..peptidoglycan.biosynthesis.I..meso.diaminopimelate.containing."
## [37] "PHOSLIPSYN.PWY..superpathway.of.phospholipid.biosynthesis.I..bacteria."
## [38] "POLYISOPRENSYN.PWY..polyisoprenoid.biosynthesis..E..coli."
## [39] "PWY.1042..glycolysis.IV..plant.cytosol."
## [40] "PWY.1269..CMP.3.deoxy.D.manno.octulosonate.biosynthesis.I"
## [41] "PWY.2941..L.lysine.biosynthesis.II"
## [42] "PWY.2942..L.lysine.biosynthesis.III"
## [43] "PWY.3001..superpathway.of.L.isoleucine.biosynthesis.I"
## [44] "PWY.3841..folate.transformations.II"
## [45] "PWY.4242..pantothenate.and.coenzyme.A.biosynthesis.III"
## [46] "PWY.4981..L.proline.biosynthesis.II..from.arginine."
## [47] "PWY.4984..urea.cycle"
## [48] "PWY.5030..L.histidine.degradation.III"
## [49] "PWY.5097..L.lysine.biosynthesis.VI"
## [50] "PWY.5103..L.isoleucine.biosynthesis.III"
## [51] "PWY.5154..L.arginine.biosynthesis.III..via.N.acetyl.L.citrulline."
## [52] "PWY.5304..superpathway.of.sulfur.oxidation..Acidianus.ambivalens."
## [53] "PWY.5484..glycolysis.II..from.fructose.6.phosphate."
## [54] "PWY.5505..L.glutamate.and.L.glutamine.biosynthesis"
## [55] "PWY.5659..GDP.mannose.biosynthesis"
## [56] "PWY.5667..CDP.diacylglycerol.biosynthesis.I"
## [57] "PWY.5686..UMP.biosynthesis"
## [58] "PWY.5695..urate.biosynthesis.inosine.5..phosphate.degradation"
## [59] "PWY.5941..glycogen.degradation.II..eukaryotic."
## [60] "PWY.5973..cis.vaccenate.biosynthesis"
## [61] "PWY.6121..5.aminoimidazole.ribonucleotide.biosynthesis.I"
## [62] "PWY.6122..5.aminoimidazole.ribonucleotide.biosynthesis.II"
## [63] "PWY.6123..inosine.5..phosphate.biosynthesis.I"
## [64] "PWY.6124..inosine.5..phosphate.biosynthesis.II"
## [65] "PWY.6125..superpathway.of.guanosine.nucleotides.de.novo.biosynthesis.II"
## [66] "PWY.6126..superpathway.of.adenosine.nucleotides.de.novo.biosynthesis.II"
## [67] "PWY.6147..6.hydroxymethyl.dihydropterin.diphosphate.biosynthesis.I"
## [68] "PWY.6151..S.adenosyl.L.methionine.cycle.I"
## [69] "PWY.6163..chorismate.biosynthesis.from.3.dehydroquinate"
## [70] "PWY.6168..flavin.biosynthesis.III..fungi."
## [71] "PWY.621..sucrose.degradation.III..sucrose.invertase."
## [72] "PWY.6277..superpathway.of.5.aminoimidazole.ribonucleotide.biosynthesis"
## [73] "PWY.6305..putrescine.biosynthesis.IV"
## [74] "PWY.6317..galactose.degradation.I..Leloir.pathway."
## [75] "PWY.6385..peptidoglycan.biosynthesis.III..mycobacteria."
## [76] "PWY.6386..UDP.N.acetylmuramoyl.pentapeptide.biosynthesis.II..lysine.containing."
## [77] "PWY.6387..UDP.N.acetylmuramoyl.pentapeptide.biosynthesis.I..meso.diaminopimelate.containing."
## [78] "PWY.6507..4.deoxy.L.threo.hex.4.enopyranuronate.degradation"
## [79] "PWY.6545..pyrimidine.deoxyribonucleotides.de.novo.biosynthesis.III"
## [80] "PWY.6572..chondroitin.sulfate.degradation.I..bacterial."
## [81] "PWY.6609..adenine.and.adenosine.salvage.III"
## [82] "PWY.6700..queuosine.biosynthesis"
## [83] "PWY.6703..preQ0.biosynthesis"
## [84] "PWY.6737..starch.degradation.V"
## [85] "PWY.6859..all.trans.farnesol.biosynthesis"
## [86] "PWY.6892..thiazole.biosynthesis.I..E..coli."
## [87] "PWY.6897..thiamin.salvage.II"
## [88] "PWY.6901..superpathway.of.glucose.and.xylose.degradation"
## [89] "PWY.6936..seleno.amino.acid.biosynthesis"
## [90] "PWY.7111..pyruvate.fermentation.to.isobutanol..engineered."
## [91] "PWY.7184..pyrimidine.deoxyribonucleotides.de.novo.biosynthesis.I"
## [92] "PWY.7187..pyrimidine.deoxyribonucleotides.de.novo.biosynthesis.II"
## [93] "PWY.7197..pyrimidine.deoxyribonucleotide.phosphorylation"
## [94] "PWY.7199..pyrimidine.deoxyribonucleosides.salvage"
## [95] "PWY.7208..superpathway.of.pyrimidine.nucleobases.salvage"
## [96] "PWY.7210..pyrimidine.deoxyribonucleotides.biosynthesis.from.CTP"
## [97] "PWY.7211..superpathway.of.pyrimidine.deoxyribonucleotides.de.novo.biosynthesis"
## [98] "PWY.7219..adenosine.ribonucleotides.de.novo.biosynthesis"
## [99] "PWY.7220..adenosine.deoxyribonucleotides.de.novo.biosynthesis.II"
## [100] "PWY.7221..guanosine.ribonucleotides.de.novo.biosynthesis"
## [101] "PWY.7222..guanosine.deoxyribonucleotides.de.novo.biosynthesis.II"
## [102] "PWY.7228..superpathway.of.guanosine.nucleotides.de.novo.biosynthesis.I"
## [103] "PWY.7229..superpathway.of.adenosine.nucleotides.de.novo.biosynthesis.I"
## [104] "PWY.7234..inosine.5..phosphate.biosynthesis.III"
## [105] "PWY.724..superpathway.of.L.lysine..L.threonine.and.L.methionine.biosynthesis.II"
## [106] "PWY.7242..D.fructuronate.degradation"
## [107] "PWY.7282..4.amino.2.methyl.5.phosphomethylpyrimidine.biosynthesis..yeast."
## [108] "PWY.7323..superpathway.of.GDP.mannose.derived.O.antigen.building.blocks.biosynthesis"
## [109] "PWY.7332..superpathway.of.UDP.N.acetylglucosamine.derived.O.antigen.building.blocks.biosynthesis"
## [110] "PWY.7357..thiamin.formation.from.pyrithiamine.and.oxythiamine..yeast."
## [111] "PWY.7383..anaerobic.energy.metabolism..invertebrates..cytosol."
## [112] "PWY.7400..L.arginine.biosynthesis.IV..archaebacteria."
## [113] "PWY.7456..mannan.degradation"
## [114] "PWY.7539..6.hydroxymethyl.dihydropterin.diphosphate.biosynthesis.III..Chlamydia."
## [115] "PWY.7663..gondoate.biosynthesis..anaerobic."
## [116] "PWY.841..superpathway.of.purine.nucleotides.de.novo.biosynthesis.I"
## [117] "PWY0.1296..purine.ribonucleosides.degradation"
## [118] "PWY0.1319..CDP.diacylglycerol.biosynthesis.II"
## [119] "PWY0.1586..peptidoglycan.maturation..meso.diaminopimelate.containing."
## [120] "PWY0.162..superpathway.of.pyrimidine.ribonucleotides.de.novo.biosynthesis"
## [121] "PWY0.166..superpathway.of.pyrimidine.deoxyribonucleotides.de.novo.biosynthesis..E..coli."
## [122] "PWY0.845..superpathway.of.pyridoxal.5..phosphate.biosynthesis.and.salvage"
## [123] "PWY4FS.7..phosphatidylglycerol.biosynthesis.I..plastidic."
## [124] "PWY4FS.8..phosphatidylglycerol.biosynthesis.II..non.plastidic."
## [125] "PWY66.399..gluconeogenesis.III"
## [126] "PWY66.422..D.galactose.degradation.V..Leloir.pathway."
## [127] "PYRIDNUCSYN.PWY..NAD.biosynthesis.I..from.aspartate."
## [128] "PYRIDOXSYN.PWY..pyridoxal.5..phosphate.biosynthesis.I"
## [129] "RHAMCAT.PWY..L.rhamnose.degradation.I"
## [130] "RIBOSYN2.PWY..flavin.biosynthesis.I..bacteria.and.plants."
## [131] "SER.GLYSYN.PWY..superpathway.of.L.serine.and.glycine.biosynthesis.I"
## [132] "TEICHOICACID.PWY..teichoic.acid..poly.glycerol..biosynthesis"
## [133] "THISYN.PWY..superpathway.of.thiamin.diphosphate.biosynthesis.I"
## [134] "THISYNARA.PWY..superpathway.of.thiamin.diphosphate.biosynthesis.III..eukaryotes."
## [135] "THRESYN.PWY..superpathway.of.L.threonine.biosynthesis"
## [136] "TRNA.CHARGING.PWY..tRNA.charging"
## [137] "TRPSYN.PWY..L.tryptophan.biosynthesis"
## [138] "VALSYN.PWY..L.valine.biosynthesis"
## [139] "X1CMET2.PWY..N10.formyl.tetrahydrofolate.biosynthesis"
##
## $MEbrown
## [1] "GLCMANNANAUT.PWY..superpathway.of.N.acetylglucosamine..N.acetylmannosamine.and.N.acetylneuraminate.degradation"
## [2] "GLYCOLYSIS.E.D..superpathway.of.glycolysis.and.Entner.Doudoroff"
## [3] "METH.ACETATE.PWY..methanogenesis.from.acetate"
## [4] "PWY.5100..pyruvate.fermentation.to.acetate.and.lactate.II"
## [5] "PWY.5104..L.isoleucine.biosynthesis.IV"
## [6] "PWY.5177..glutaryl.CoA.degradation"
## [7] "PWY.5690..TCA.cycle.II..plants.and.fungi."
## [8] "PWY.6470..peptidoglycan.biosynthesis.V...beta..lactam.resistance."
## [9] "PWY.6527..stachyose.degradation"
## [10] "PWY.6595..superpathway.of.guanosine.nucleotides.degradation..plants."
## [11] "PWY.6608..guanosine.nucleotides.degradation.III"
## [12] "PWY.7196..superpathway.of.pyrimidine.ribonucleosides.salvage"
## [13] "PWY.7198..pyrimidine.deoxyribonucleotides.de.novo.biosynthesis.IV"
## [14] "PWY0.1061..superpathway.of.L.alanine.biosynthesis"
## [15] "PWY0.1261..anhydromuropeptides.recycling"
## [16] "PWY66.400..glycolysis.VI..metazoan."
## [17] "TCA..TCA.cycle.I..prokaryotic."
## [18] "UDPNAGSYN.PWY..UDP.N.acetyl.D.glucosamine.biosynthesis.I"
##
## $MEgrey
## [1] "NAD.BIOSYNTHESIS.II..NAD.salvage.pathway.II"
## [2] "P162.PWY..L.glutamate.degradation.V..via.hydroxyglutarate."
## [3] "P163.PWY..L.lysine.fermentation.to.acetate.and.butanoate"
## [4] "PWY.3781..aerobic.respiration.I..cytochrome.c."
## [5] "PWY.6143..CMP.pseudaminate.biosynthesis"
## [6] "PWY.622..starch.biosynthesis"
## [7] "PWY.6263..superpathway.of.menaquinol.8.biosynthesis.II"
## [8] "PWY.6478..GDP.D.glycero..alpha..D.manno.heptose.biosynthesis"
## [9] "PWY.6749..CMP.legionaminate.biosynthesis.I"
## [10] "PWY.6953..dTDP.3.acetamido.3.6.dideoxy..alpha..D.galactose.biosynthesis"
## [11] "PWY.7237..myo...chiro..and.scillo.inositol.degradation"
## [12] "PWY.7312..dTDP.D..beta..fucofuranose.biosynthesis"
## [13] "PWY.7316..dTDP.N.acetylviosamine.biosynthesis"
## [14] "PWY.7371..1.4.dihydroxy.6.naphthoate.biosynthesis.II"
## [15] "PWY490.3..nitrate.reduction.VI..assimilatory."
## [16] "RUMP.PWY..formaldehyde.oxidation.I"
##
## $MEturquoise
## [1] "ALLANTOINDEG.PWY..superpathway.of.allantoin.degradation.in.yeast"
## [2] "ARG.POLYAMINE.SYN..superpathway.of.arginine.and.polyamine.biosynthesis"
## [3] "ARGDEG.PWY..superpathway.of.L.arginine..putrescine..and.4.aminobutanoate.degradation"
## [4] "ARGORNPROST.PWY..arginine..ornithine.and.proline.interconversion"
## [5] "AST.PWY..L.arginine.degradation.II..AST.pathway."
## [6] "BIOTIN.BIOSYNTHESIS.PWY..biotin.biosynthesis.I"
## [7] "CENTFERM.PWY..pyruvate.fermentation.to.butanoate"
## [8] "DAPLYSINESYN.PWY..L.lysine.biosynthesis.I"
## [9] "ECASYN.PWY..enterobacterial.common.antigen.biosynthesis"
## [10] "ENTBACSYN.PWY..enterobactin.biosynthesis"
## [11] "FAO.PWY..fatty.acid..beta..oxidation.I"
## [12] "FASYN.ELONG.PWY..fatty.acid.elongation....saturated"
## [13] "FASYN.INITIAL.PWY..superpathway.of.fatty.acid.biosynthesis.initiation..E..coli."
## [14] "FERMENTATION.PWY..mixed.acid.fermentation"
## [15] "FOLSYN.PWY..superpathway.of.tetrahydrofolate.biosynthesis.and.salvage"
## [16] "FUC.RHAMCAT.PWY..superpathway.of.fucose.and.rhamnose.degradation"
## [17] "FUCCAT.PWY..fucose.degradation"
## [18] "GALACTARDEG.PWY..D.galactarate.degradation.I"
## [19] "GLUCARDEG.PWY..D.glucarate.degradation.I"
## [20] "GLUCARGALACTSUPER.PWY..superpathway.of.D.glucarate.and.D.galactarate.degradation"
## [21] "GLUCOSE1PMETAB.PWY..glucose.and.glucose.1.phosphate.degradation"
## [22] "GLUDEG.I.PWY..GABA.shunt"
## [23] "GLYCOCAT.PWY..glycogen.degradation.I..bacterial."
## [24] "GLYCOL.GLYOXDEG.PWY..superpathway.of.glycol.metabolism.and.degradation"
## [25] "GLYCOLYSIS.TCA.GLYOX.BYPASS..superpathway.of.glycolysis..pyruvate.dehydrogenase..TCA..and.glyoxylate.bypass"
## [26] "GLYOXYLATE.BYPASS..glyoxylate.cycle"
## [27] "GOLPDLCAT.PWY..superpathway.of.glycerol.degradation.to.1.3.propanediol"
## [28] "HCAMHPDEG.PWY..3.phenylpropanoate.and.3..3.hydroxyphenyl.propanoate.degradation.to.2.oxopent.4.enoate"
## [29] "HEME.BIOSYNTHESIS.II..heme.biosynthesis.I..aerobic."
## [30] "HEMESYN2.PWY..heme.biosynthesis.II..anaerobic."
## [31] "HEXITOLDEGSUPER.PWY..superpathway.of.hexitol.degradation..bacteria."
## [32] "HOMOSER.METSYN.PWY..L.methionine.biosynthesis.I"
## [33] "KDO.NAGLIPASYN.PWY..superpathway.of..Kdo.2.lipid.A.biosynthesis"
## [34] "KETOGLUCONMET.PWY..ketogluconate.metabolism"
## [35] "LACTOSECAT.PWY..lactose.and.galactose.degradation.I"
## [36] "LPSSYN.PWY..superpathway.of.lipopolysaccharide.biosynthesis"
## [37] "MET.SAM.PWY..superpathway.of.S.adenosyl.L.methionine.biosynthesis"
## [38] "METHGLYUT.PWY..superpathway.of.methylglyoxal.degradation"
## [39] "METSYN.PWY..L.homoserine.and.L.methionine.biosynthesis"
## [40] "NONMEVIPP.PWY..methylerythritol.phosphate.pathway.I"
## [41] "ORNARGDEG.PWY..superpathway.of.L.arginine.and.L.ornithine.degradation"
## [42] "ORNDEG.PWY..superpathway.of.ornithine.degradation"
## [43] "P105.PWY..TCA.cycle.IV..2.oxoglutarate.decarboxylase."
## [44] "P122.PWY..heterolactic.fermentation"
## [45] "P124.PWY..Bifidobacterium.shunt"
## [46] "P161.PWY..acetylene.degradation"
## [47] "P164.PWY..purine.nucleobases.degradation.I..anaerobic."
## [48] "P185.PWY..formaldehyde.assimilation.III..dihydroxyacetone.cycle."
## [49] "P221.PWY..octane.oxidation"
## [50] "P23.PWY..reductive.TCA.cycle.I"
## [51] "P4.PWY..superpathway.of.L.lysine..L.threonine.and.L.methionine.biosynthesis.I"
## [52] "P42.PWY..incomplete.reductive.TCA.cycle"
## [53] "P441.PWY..superpathway.of.N.acetylneuraminate.degradation"
## [54] "P461.PWY..hexitol.fermentation.to.lactate..formate..ethanol.and.acetate"
## [55] "POLYAMINSYN3.PWY..superpathway.of.polyamine.biosynthesis.II"
## [56] "POLYAMSYN.PWY..superpathway.of.polyamine.biosynthesis.I"
## [57] "PPGPPMET.PWY..ppGpp.biosynthesis"
## [58] "PROTOCATECHUATE.ORTHO.CLEAVAGE.PWY..protocatechuate.degradation.II..ortho.cleavage.pathway."
## [59] "PRPP.PWY..superpathway.of.histidine..purine..and.pyrimidine.biosynthesis"
## [60] "PWY.1861..formaldehyde.assimilation.II..RuMP.Cycle."
## [61] "PWY.241..C4.photosynthetic.carbon.assimilation.cycle..NADP.ME.type"
## [62] "PWY.2723..trehalose.degradation.V"
## [63] "PWY.4041...gamma..glutamyl.cycle"
## [64] "PWY.4702..phytate.degradation.I"
## [65] "PWY.5005..biotin.biosynthesis.II"
## [66] "PWY.5022..4.aminobutanoate.degradation.V"
## [67] "PWY.5083..NAD.NADH.phosphorylation.and.dephosphorylation"
## [68] "PWY.5088..L.glutamate.degradation.VIII..to.propanoate."
## [69] "PWY.5121..superpathway.of.geranylgeranyl.diphosphate.biosynthesis.II..via.MEP."
## [70] "PWY.5136..fatty.acid..beta..oxidation.II..peroxisome."
## [71] "PWY.5138..unsaturated..even.numbered.fatty.acid..beta..oxidation"
## [72] "PWY.5173..superpathway.of.acetyl.CoA.biosynthesis"
## [73] "PWY.5188..tetrapyrrole.biosynthesis.I..from.glutamate."
## [74] "PWY.5189..tetrapyrrole.biosynthesis.II..from.glycine."
## [75] "PWY.5345..superpathway.of.L.methionine.biosynthesis..by.sulfhydrylation."
## [76] "PWY.5347..superpathway.of.L.methionine.biosynthesis..transsulfuration."
## [77] "PWY.5367..petroselinate.biosynthesis"
## [78] "PWY.5384..sucrose.degradation.IV..sucrose.phosphorylase."
## [79] "PWY.5464..superpathway.of.cytosolic.glycolysis..plants...pyruvate.dehydrogenase.and.TCA.cycle"
## [80] "PWY.561..superpathway.of.glyoxylate.cycle.and.fatty.acid.degradation"
## [81] "PWY.5656..mannosylglycerate.biosynthesis.I"
## [82] "PWY.5675..nitrate.reduction.V..assimilatory."
## [83] "PWY.5676..acetyl.CoA.fermentation.to.butanoate.II"
## [84] "PWY.5677..succinate.fermentation.to.butanoate"
## [85] "PWY.5692..allantoin.degradation.to.glyoxylate.II"
## [86] "PWY.5705..allantoin.degradation.to.glyoxylate.III"
## [87] "PWY.5723..Rubisco.shunt"
## [88] "PWY.5747..2.methylcitrate.cycle.II"
## [89] "PWY.5791..1.4.dihydroxy.2.naphthoate.biosynthesis.II..plants."
## [90] "PWY.5837..1.4.dihydroxy.2.naphthoate.biosynthesis.I"
## [91] "PWY.5838..superpathway.of.menaquinol.8.biosynthesis.I"
## [92] "PWY.5840..superpathway.of.menaquinol.7.biosynthesis"
## [93] "PWY.5845..superpathway.of.menaquinol.9.biosynthesis"
## [94] "PWY.5850..superpathway.of.menaquinol.6.biosynthesis.I"
## [95] "PWY.5855..ubiquinol.7.biosynthesis..prokaryotic."
## [96] "PWY.5856..ubiquinol.9.biosynthesis..prokaryotic."
## [97] "PWY.5857..ubiquinol.10.biosynthesis..prokaryotic."
## [98] "PWY.5860..superpathway.of.demethylmenaquinol.6.biosynthesis.I"
## [99] "PWY.5861..superpathway.of.demethylmenaquinol.8.biosynthesis"
## [100] "PWY.5862..superpathway.of.demethylmenaquinol.9.biosynthesis"
## [101] "PWY.5863..superpathway.of.phylloquinol.biosynthesis"
## [102] "PWY.5896..superpathway.of.menaquinol.10.biosynthesis"
## [103] "PWY.5897..superpathway.of.menaquinol.11.biosynthesis"
## [104] "PWY.5898..superpathway.of.menaquinol.12.biosynthesis"
## [105] "PWY.5899..superpathway.of.menaquinol.13.biosynthesis"
## [106] "PWY.5913..TCA.cycle.VI..obligate.autotrophs."
## [107] "PWY.5918..superpathay.of.heme.biosynthesis.from.glutamate"
## [108] "PWY.5920..superpathway.of.heme.biosynthesis.from.glycine"
## [109] "PWY.5971..palmitate.biosynthesis.II..bacteria.and.plants."
## [110] "PWY.5989..stearate.biosynthesis.II..bacteria.and.plants."
## [111] "PWY.6113..superpathway.of.mycolate.biosynthesis"
## [112] "PWY.6270..isoprene.biosynthesis.I"
## [113] "PWY.6282..palmitoleate.biosynthesis.I..from..5Z..dodec.5.enoate."
## [114] "PWY.6284..superpathway.of.unsaturated.fatty.acids.biosynthesis..E..coli."
## [115] "PWY.6285..superpathway.of.fatty.acids.biosynthesis..E..coli."
## [116] "PWY.6318..L.phenylalanine.degradation.IV..mammalian..via.side.chain."
## [117] "PWY.6353..purine.nucleotides.degradation.II..aerobic."
## [118] "PWY.6471..peptidoglycan.biosynthesis.IV..Enterococcus.faecium."
## [119] "PWY.6519..8.amino.7.oxononanoate.biosynthesis.I"
## [120] "PWY.6531..mannitol.cycle"
## [121] "PWY.6549..L.glutamine.biosynthesis.III"
## [122] "PWY.6588..pyruvate.fermentation.to.acetone"
## [123] "PWY.6590..superpathway.of.Clostridium.acetobutylicum.acidogenic.fermentation"
## [124] "PWY.6606..guanosine.nucleotides.degradation.II"
## [125] "PWY.6612..superpathway.of.tetrahydrofolate.biosynthesis"
## [126] "PWY.6628..superpathway.of.L.phenylalanine.biosynthesis"
## [127] "PWY.6629..superpathway.of.L.tryptophan.biosynthesis"
## [128] "PWY.6630..superpathway.of.L.tyrosine.biosynthesis"
## [129] "PWY.6690..cinnamate.and.3.hydroxycinnamate.degradation.to.2.oxopent.4.enoate"
## [130] "PWY.6708..ubiquinol.8.biosynthesis..prokaryotic."
## [131] "PWY.6731..starch.degradation.III"
## [132] "PWY.6803..phosphatidylcholine.acyl.editing"
## [133] "PWY.6823..molybdenum.cofactor.biosynthesis"
## [134] "PWY.6837..fatty.acid.beta.oxidation.V..unsaturated..odd.number..di.isomerase.dependent."
## [135] "PWY.6876..isopropanol.biosynthesis"
## [136] "PWY.6891..thiazole.biosynthesis.II..Bacillus."
## [137] "PWY.6895..superpathway.of.thiamin.diphosphate.biosynthesis.II"
## [138] "PWY.6969..TCA.cycle.V..2.oxoglutarate.ferredoxin.oxidoreductase."
## [139] "PWY.7003..glycerol.degradation.to.butanol"
## [140] "PWY.7013..L.1.2.propanediol.degradation"
## [141] "PWY.7046..4.coumarate.degradation..anaerobic."
## [142] "PWY.7094..fatty.acid.salvage"
## [143] "PWY.7115..C4.photosynthetic.carbon.assimilation.cycle..NAD.ME.type"
## [144] "PWY.7117..C4.photosynthetic.carbon.assimilation.cycle..PEPCK.type"
## [145] "PWY.7204..pyridoxal.5..phosphate.salvage.II..plants."
## [146] "PWY.7209..superpathway.of.pyrimidine.ribonucleosides.degradation"
## [147] "PWY.7254..TCA.cycle.VII..acetate.producers."
## [148] "PWY.7269..NAD.NADP.NADH.NADPH.mitochondrial.interconversion..yeast."
## [149] "PWY.7279..aerobic.respiration.II..cytochrome.c...yeast."
## [150] "PWY.7288..fatty.acid..beta..oxidation..peroxisome..yeast."
## [151] "PWY.7315..dTDP.N.acetylthomosamine.biosynthesis"
## [152] "PWY.7328..superpathway.of.UDP.glucose.derived.O.antigen.building.blocks.biosynthesis"
## [153] "PWY.7385..1.3.propanediol.biosynthesis..engineered."
## [154] "PWY.7388..octanoyl..acyl.carrier.protein..biosynthesis..mitochondria..yeast."
## [155] "PWY.7392..taxadiene.biosynthesis..engineered."
## [156] "PWY.7446..sulfoglycolysis"
## [157] "PWY.7560..methylerythritol.phosphate.pathway.II"
## [158] "PWY.7616..methanol.oxidation.to.carbon.dioxide"
## [159] "PWY.7664..oleate.biosynthesis.IV..anaerobic."
## [160] "PWY.821..superpathway.of.sulfur.amino.acid.biosynthesis..Saccharomyces.cerevisiae."
## [161] "PWY0.1241..ADP.L.glycero..beta..D.manno.heptose.biosynthesis"
## [162] "PWY0.1277..3.phenylpropanoate.and.3..3.hydroxyphenyl.propanoate.degradation"
## [163] "PWY0.1297..superpathway.of.purine.deoxyribonucleosides.degradation"
## [164] "PWY0.1298..superpathway.of.pyrimidine.deoxyribonucleosides.degradation"
## [165] "PWY0.1338..polymyxin.resistance"
## [166] "PWY0.1415..superpathway.of.heme.biosynthesis.from.uroporphyrinogen.III"
## [167] "PWY0.1479..tRNA.processing"
## [168] "PWY0.1533..methylphosphonate.degradation.I"
## [169] "PWY0.41..allantoin.degradation.IV..anaerobic."
## [170] "PWY0.42..2.methylcitrate.cycle.I"
## [171] "PWY0.781..aspartate.superpathway"
## [172] "PWY0.862...5Z..dodec.5.enoate.biosynthesis"
## [173] "PWY0.881..superpathway.of.fatty.acid.biosynthesis.I..E..coli."
## [174] "PWY3O.355..stearate.biosynthesis.III..fungi."
## [175] "PWY4LZ.257..superpathway.of.fermentation..Chlamydomonas.reinhardtii."
## [176] "PWY66.389..phytol.degradation"
## [177] "PWY66.391..fatty.acid..beta..oxidation.VI..peroxisome."
## [178] "PWY66.398..TCA.cycle.III..animals."
## [179] "PWY66.409..superpathway.of.purine.nucleotide.salvage"
## [180] "PWYG.321..mycolate.biosynthesis"
## [181] "PYRIDNUCSAL.PWY..NAD.salvage.pathway.I"
## [182] "REDCITCYC..TCA.cycle.VIII..helicobacter."
## [183] "SALVADEHYPOX.PWY..adenosine.nucleotides.degradation.II"
## [184] "SO4ASSIM.PWY..sulfate.reduction.I..assimilatory."
## [185] "SULFATE.CYS.PWY..superpathway.of.sulfate.assimilation.and.cysteine.biosynthesis"
## [186] "TCA.GLYOX.BYPASS..superpathway.of.glyoxylate.bypass.and.TCA"
## [187] "THREOCAT.PWY..superpathway.of.L.threonine.metabolism"
## [188] "UBISYN.PWY..superpathway.of.ubiquinol.8.biosynthesis..prokaryotic."
## [189] "URDEGR.PWY..superpathway.of.allantoin.degradation.in.plants"
## [190] "X3.HYDROXYPHENYLACETATE.DEGRADATION.PWY..4.hydroxyphenylacetate.degradation"
############
# Run MSEA #
############
MSEA <- run_MSEA(microbeSet, results$feature)
MSEA <- MSEA[, c('Set', 'Freq', 'ES', setdiff(names(MSEA), c('Set', 'Freq', 'ES')))]
colnames(MSEA) <- c('ID', 'Size', 'pval', 'qval')
MSEA$ID <- paste(MSEA$ID, ' (', MSEA$Size, ')', sep = '')
########
# Plot #
########
p <- MSEA %>%
arrange(-pval) %>%
mutate(ID = factor(ID, levels = ID)) %>%
ggplot(aes(y = -log10(pval), x = ID)) +
geom_bar(stat = "identity", fill = 'cornflowerblue') + theme_bw() +
coord_flip() +
ggtitle('Statistically significant modules associated with disease') +
xlab('') +
ylab('MSEA enrichment score')
print(p)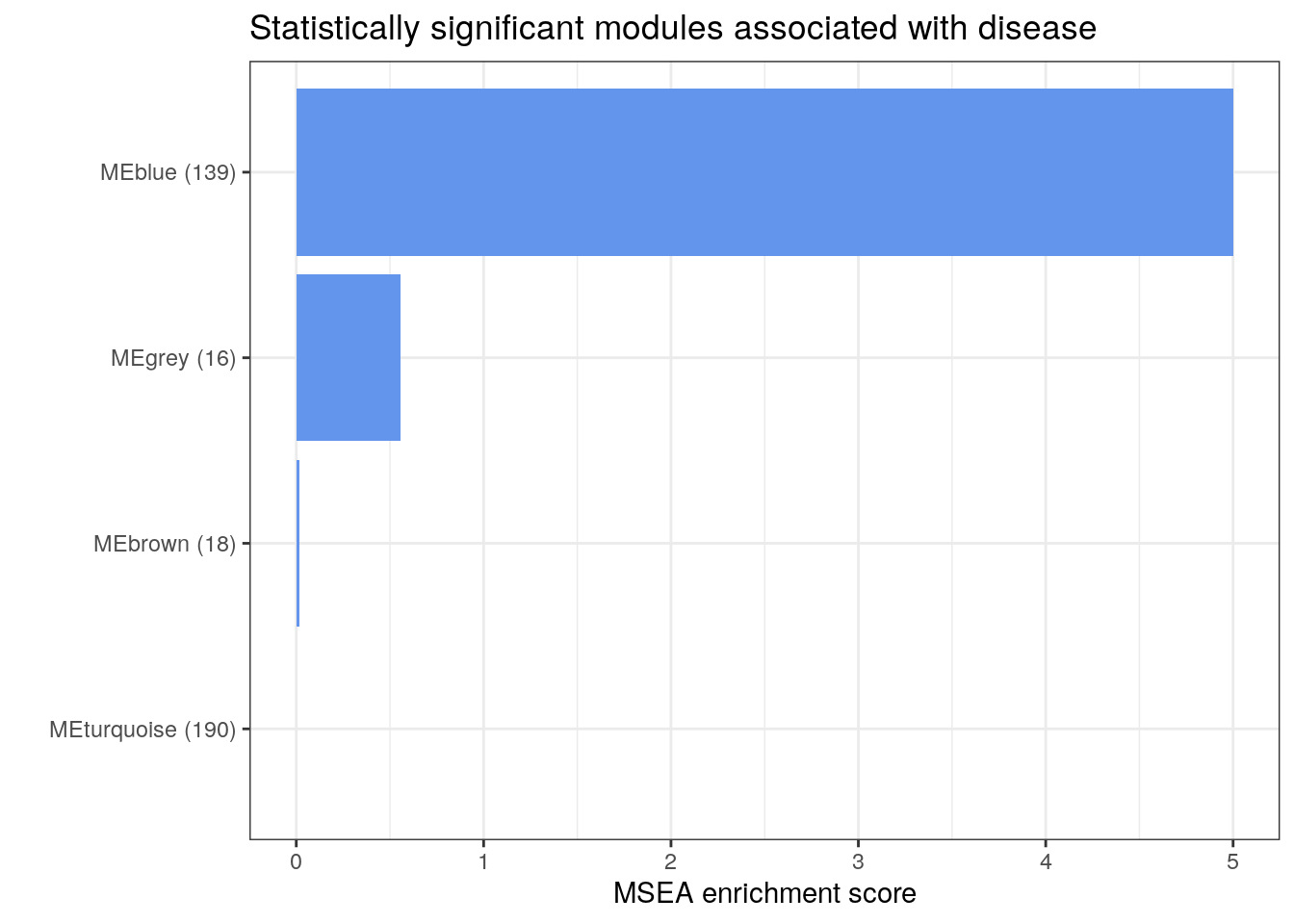
Based on the MSEA results, we obtain 4 enriched modules of microbial pathways. We can similarly examine the members of the top enriched modules.
17.5 Print the most significant modules for pathway abundance data
# Print the most significant modules
microbeSet[["MEpurple"]]
## NULL
microbeSet[["MEbrown"]]
## [1] "GLCMANNANAUT.PWY..superpathway.of.N.acetylglucosamine..N.acetylmannosamine.and.N.acetylneuraminate.degradation"
## [2] "GLYCOLYSIS.E.D..superpathway.of.glycolysis.and.Entner.Doudoroff"
## [3] "METH.ACETATE.PWY..methanogenesis.from.acetate"
## [4] "PWY.5100..pyruvate.fermentation.to.acetate.and.lactate.II"
## [5] "PWY.5104..L.isoleucine.biosynthesis.IV"
## [6] "PWY.5177..glutaryl.CoA.degradation"
## [7] "PWY.5690..TCA.cycle.II..plants.and.fungi."
## [8] "PWY.6470..peptidoglycan.biosynthesis.V...beta..lactam.resistance."
## [9] "PWY.6527..stachyose.degradation"
## [10] "PWY.6595..superpathway.of.guanosine.nucleotides.degradation..plants."
## [11] "PWY.6608..guanosine.nucleotides.degradation.III"
## [12] "PWY.7196..superpathway.of.pyrimidine.ribonucleosides.salvage"
## [13] "PWY.7198..pyrimidine.deoxyribonucleotides.de.novo.biosynthesis.IV"
## [14] "PWY0.1061..superpathway.of.L.alanine.biosynthesis"
## [15] "PWY0.1261..anhydromuropeptides.recycling"
## [16] "PWY66.400..glycolysis.VI..metazoan."
## [17] "TCA..TCA.cycle.I..prokaryotic."
## [18] "UDPNAGSYN.PWY..UDP.N.acetyl.D.glucosamine.biosynthesis.I"