1 Microbiome data science in Bioconductor
This work - Orchestrating Microbiome Analysis with Bioconductor (Tuomas Borman et al. 2024) - contributes novel methods and educational resources for microbiome data science. It aims to teach the grammar of Bioconductor workflows in the context of microbiome data science. We show, through concrete examples, how to use the latest developments and data analytical strategies in R/Bioconductor for the manipulation, analysis, and reproducible reporting of hierarchical, heterogeneous, and multi-modal microbiome profiling data. The data science methodology is tightly integrated with the broader R/Bioconductor ecosystem. The support for modularity and interoperability is key to efficient resource sharing and collaborative development both within and across research fields.
1.1 Bioconductor
Bioconductor is a project that focuses on the development of high-quality open research software for life sciences (Gentleman et al. 2004; Huber et al. 2015). The software packages are primarily coded in R, and they undergo continuous testing and peer review to ensure high quality.
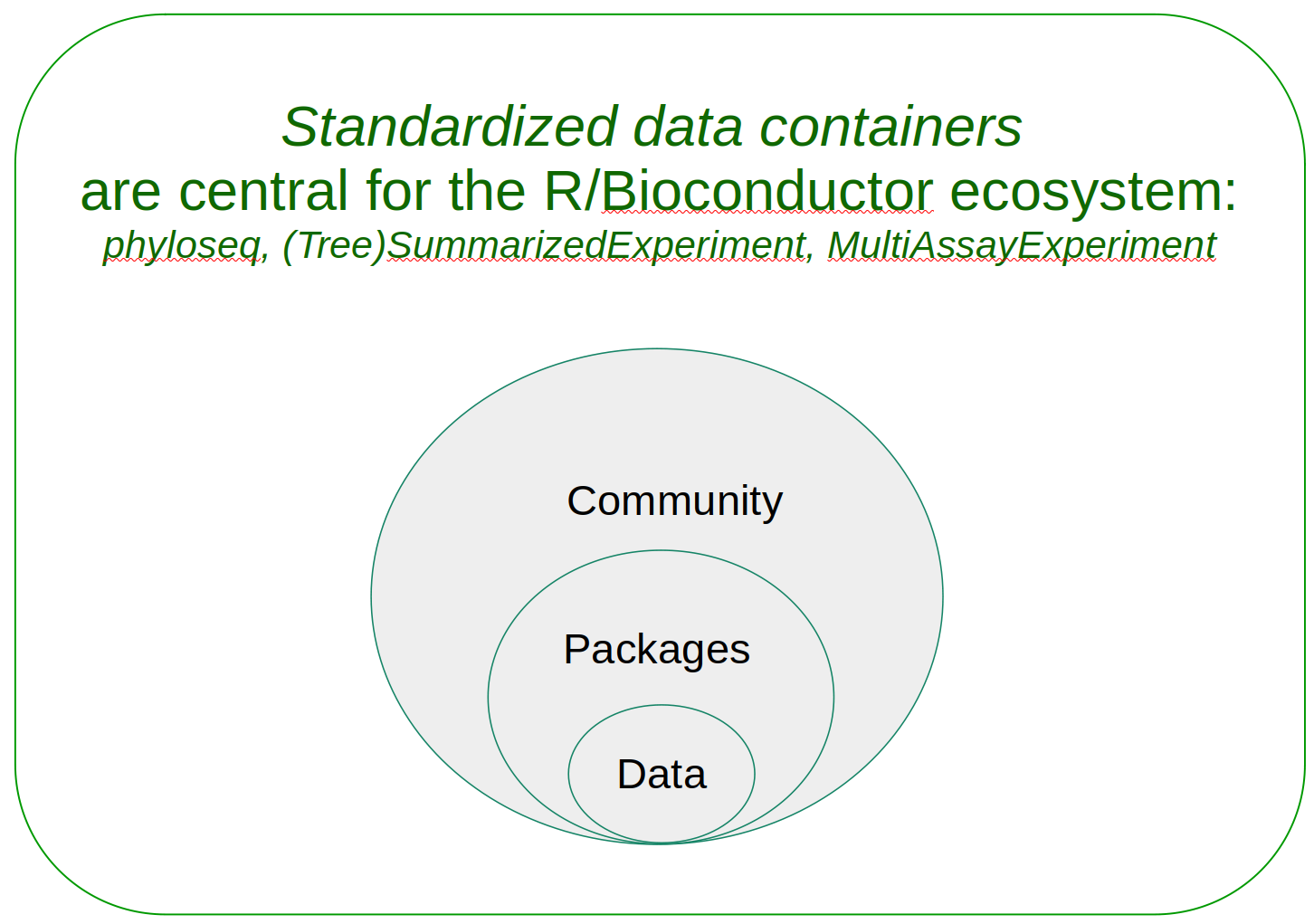
Central to the software in Bioconductor are data containers, which provide a structured presentation of data. A data container consists of slots that are dedicated to certain type of data, for example, to abundance table and sample metadata. Biological data is often complex and multidimensional, making data containers particularly beneficial. There are several key advantages to using data containers:
- Ease of handling: Data subsetting and bookkeeping become more straightforward.
- Development efficiency: Developers can create efficient methods, knowing the data will be in a consistent format.
- User accessibility: Users can easily apply complex methods to their data.
The most common data container in Bioconductor is SummarizedExperiment. It is further expanded to fulfill needs of certain application field. SummarizedExperiment and its derivatives, have already been widely adopted in microbiome research, single cell sequencing, and in other fields, allowing rapid adoption and the extension of emerging data science techniques across application domains. See Section 2.1 for more details on how to handle data containers from the SummarizedExperiment family.
The Bioconductor microbiome data science framework consists of:
- Data containers, designed to organize multi-assay microbiome data
- R/Bioconductor packages that provide dedicated methods
- Community of users and developers
1.2 Microbiome data science in Bioconductor
While microbiota is used to refer micro-organisms within well-specified area, microbiome means microbiota and their genetic material (Marchesi and Ravel 2015). Because the complex nature of the microbiome data, computational methods are essential in microbiome research.
The phyloseq data container has been dominant in the microbiome field within Bioconductor over the past decade (McMurdie and Holmes 2013). However, the data container no longer adequately meets the needs of current research, as it was originally developed for 16S sequencing data. For instance, incorporating diverse multi-omics datasets has become increasingly common.
An optimal data container should efficiently store and manage large volumes of data, including modified or transformed copies of different measurements. Furthermore, it should seamlessly integrate into the broader ecosystem of Bioconductor, minimizing duplication of effort and facilitating interoperability with other tools and packages.
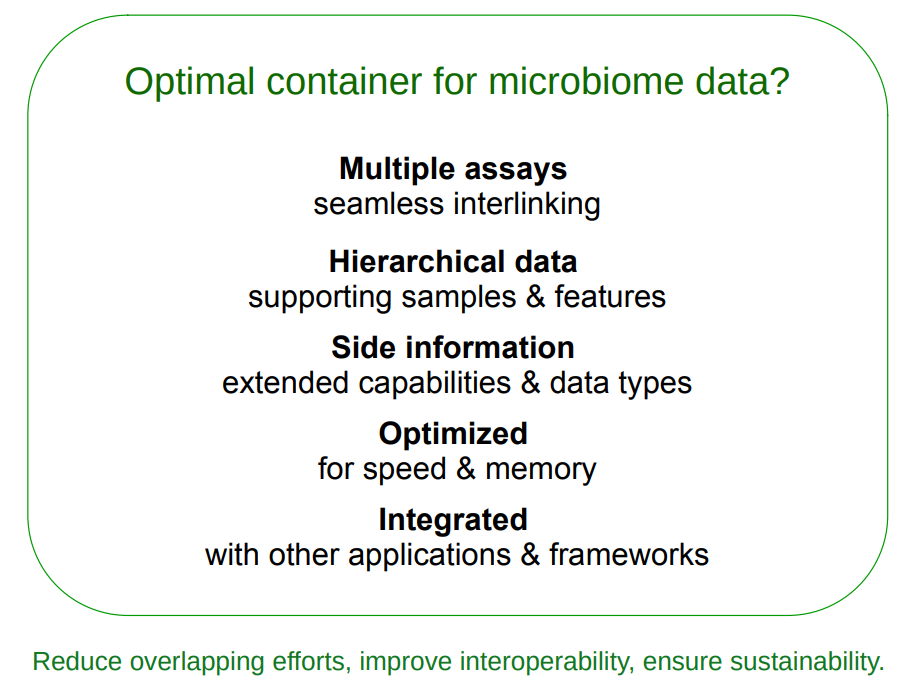
TreeSummarizedExperiment was developed to address these requirements (Huang et al. 2021). The miaverse framework was subsequently built around the TreeSummarizedExperiment data container Chapter 2.
1.3 Open data science
Open data science emphasizes sharing code and, where feasible, data alongside results (Shetty and Lahti 2019). Utilizing Bioconductor tools facilitates the development of efficient and reproducible data science workflows. Enhanced transparency in research accelerates scientific progress. As open science is a fundamental concept in microbiome research, this book aims to educate readers about reproducible reporting practices.
- Bioconductor is a large ecosystem for bioinformatics.
- Data containers are fundamental in Bioconductor.
- SummarizedExperiment is the most common data container in Bioconductor.